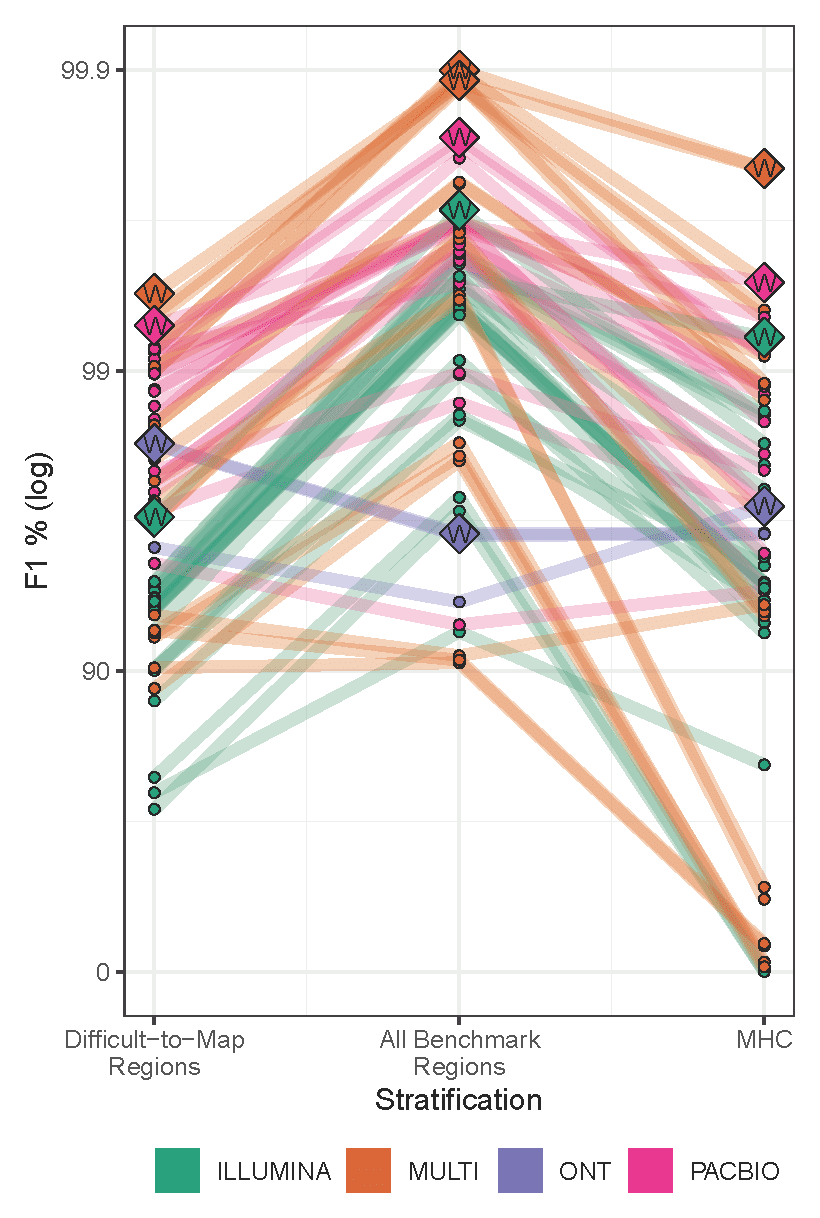
In the recent precisionFDA Truth Challenge V2, which evaluated methods for variant calling in human genomes, approaches that use PacBio HiFi reads delivered the highest precision and recall in all categories: genome-wide, specifically in difficult-to-map regions, and in the major histocompatibility complex (Figure 1). The challenge had 64 total entries: 17 using PacBio HiFi reads, 24 using Illumina reads, 3 using Oxford Nanopore reads, and 20 using multiple technologies. Twenty-five of the 26 overall most accurate callsets used PacBio HiFi reads (12 PacBio-only, 13 multi-technology), including all of the top 12 (3 PacBio-only, 9 multi-technology).
A submission from Google DeepVariant using HiFi reads achieved the highest genome-wide accuracy of any single-technology callset, with better performance for single-nucleotide variants (SNVs) and indels and 5.8× fewer total errors than the popular combination of GATK with Illumina reads (Figure 2).
The challenge was launched to evaluate variant calling for difficult regions of the human genome. Until recently, the Genome in a Bottle (GIAB) benchmarks did not measure variant calling accuracy across the most difficult 12% of the human genome, which includes many medically relevant genes. To address this, GIAB released an expanded benchmark (v4) for one of its reference samples, HG002, that covers an additional 6.3% of the genome. GIAB then developed expanded benchmarks for two other samples, HG003 and HG004. Before those benchmarks were released, the new precisionFDA challenge was used to assess currently available variant-calling techniques.
The challenge provided short reads from the Illumina NovaSeq, PacBio HiFi reads from the Sequel II System, and long reads from the Oxford Nanopore PromethION for HG002, HG003, and HG004. Competitors were invited to submit calls for HG003 and HG004, which were then evaluated against the not-yet-released “truth” variant calls for those samples. Variant calling accuracy was measured for SNVs and indels in the full genome, in difficult-to-map regions, and in the major histocompatibility complex (MHC).
The best overall performance was achieved using HiFi reads, which are both accurate (99.8%) and long (15-20 kb). HiFi read accuracy translates into accurate variant calls, and read length improves mappability to difficult regions of the genome. Equally important were advances in variant calling software, including DeepVariant (see Google AI blog for latest release) and DNAscope, to better model the properties of HiFi reads and utilize the long-range information that HiFi reads provide.
DeepVariant with only HiFi reads achieved 99.9% precision and recall for SNVs and 99.4% precision and recall for indels. In comparison, DeepVariant with Illumina reads had 4.2× more SNV errors but 1.5× fewer indel errors. The best Oxford Nanopore callset had 3.8× more SNV errors and 58.2× more indel errors (Figure 2).
The precisionFDA contest was an important opportunity to evaluate variant-calling methods, and it demonstrates how HiFi reads provide more comprehensive and accurate variant detection. We are excited to see how researchers apply this capability to search for new disease genes and to solve rare disease cases that have gone undiagnosed by other approaches.
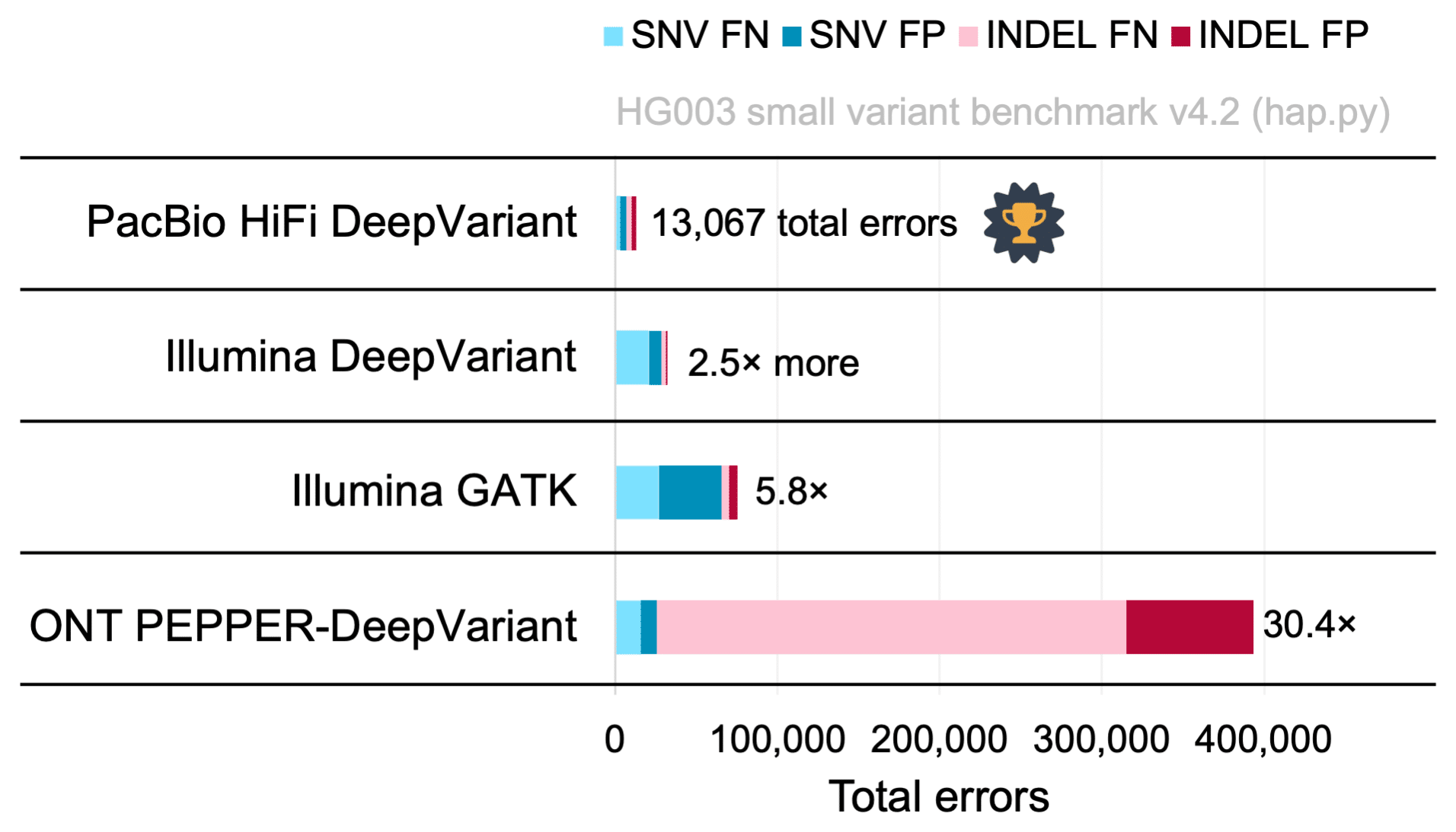 Figure 2. Variant Calling Performance, HG003. Total errors – the sum of SNV and indel false positives (FP) and false negatives (FN) – in a DeepVariant callset from 35× PacBio HiFi read coverage (Sequel II System, Chemistry 2.0); a DeepVariant callset from 35× Illumina NovaSeq; a GATK 4 callset from 35× Illumina NovaSeq; and a PEPPER-DeepVariant callset from 60× ONT PromethION R9.4.1. Performance is measured with hap.py against the Genome in a Bottle small variant benchmark v4.2 used in the precisionFDA Truth Challenge V2. The PacBio HiFi DeepVariant callset had the fewest overall errors of any single-technology callset in the challenge. Hear Aaron Wenger, a Principal Scientist at PacBio, present a summary of the results from the precisionFDA Truth Challenge V2:
Figure 2. Variant Calling Performance, HG003. Total errors – the sum of SNV and indel false positives (FP) and false negatives (FN) – in a DeepVariant callset from 35× PacBio HiFi read coverage (Sequel II System, Chemistry 2.0); a DeepVariant callset from 35× Illumina NovaSeq; a GATK 4 callset from 35× Illumina NovaSeq; and a PEPPER-DeepVariant callset from 60× ONT PromethION R9.4.1. Performance is measured with hap.py against the Genome in a Bottle small variant benchmark v4.2 used in the precisionFDA Truth Challenge V2. The PacBio HiFi DeepVariant callset had the fewest overall errors of any single-technology callset in the challenge. Hear Aaron Wenger, a Principal Scientist at PacBio, present a summary of the results from the precisionFDA Truth Challenge V2:
Explore workflows and additional resources on comprehensive variant detection or structural variant detection.