Human genomics
Uncompromising power to unlock the genome to improve human health
HUMAN GENOMICS SOLUTIONS
PacBio HiFi sequencing delivers highly accurate, single-molecule long reads for analyzing both DNA and RNA, allowing you to explore the full extent of human genetic variation with a more comprehensive view of the genome, transcriptome, and epigenome than ever before. The benefits of HiFi sequencing — and its numerous applications in human genomics — are ready to enable your next great discovery.
Genetic testing
Illuminate complex regions of the genome that are crucial for disease identification.
Clinical research
Get detailed information into complex genetic regions by detecting structural variants and repeat expansions to accelerate personalized medicine in clinical research.
Population genetics
Utilize scalable long-read sequencing to explore genomic variation, driving precision medicine initiatives in diverse populations.
Human genomics — how PacBio HiFi sequencing compares
Brochure
Human genomics
The length and accuracy of HiFi reads allow you to access genomic dark regions such as areas of high homology, GC-rich and repetitive regions, and to resolve complex gene families like HLA and pharmacogenes like CYP2D6. PacBio HiFi reads are typically around 15 kb in length with an accuracy of 99.9% and can sequence through GC-rich regions and even repeats in segmental duplications and centromeres — all while delivering simultaneous 5-methylcytosine (5mC) calls for a commanding view of the genome.
HUMAN GENOMICS IN ACTION
HUMAN GENOMICS IN ACTION
REPORT
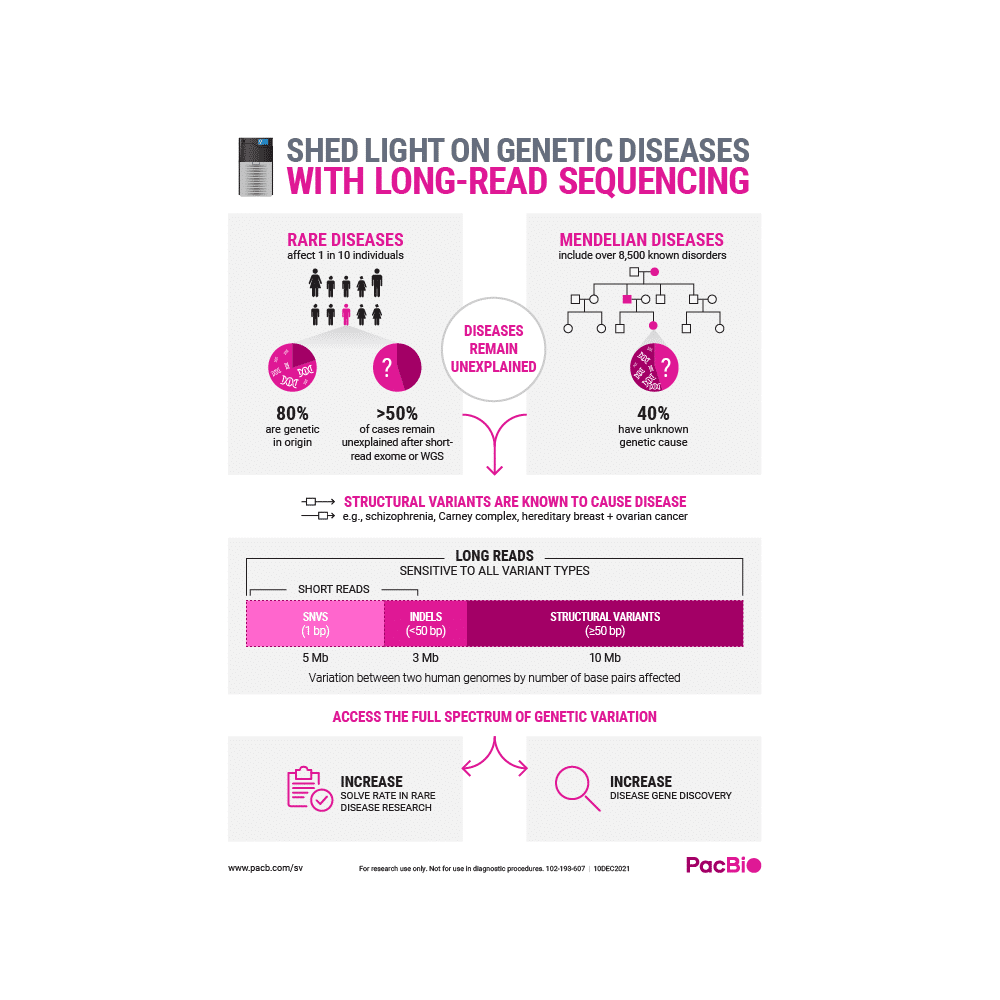
REVEAL MORE GENETIC DISEASES WITH LONG-READ SEQUENCING
REVEAL MORE GENETIC DISEASES WITH LONG-READ SEQUENCING
Identify the potential cause of unsolved genetic diseases with highly accurate HiFi sequencing.
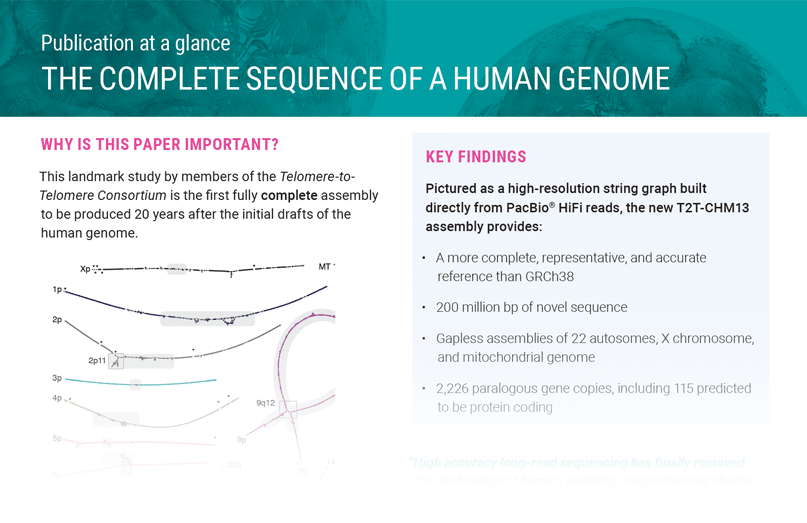
THE COMPLETE SEQUENCE OF A HUMAN GENOME
A landmark study by members of the Telomere-to-Telomere Consortium is the…

Repeat expansion panel for clinical research
The PureTarget repeat expansion panel offers a robust, end-to-end workflow designed to reduce iterative testing using legacy technology and reduce the time needed to identify neurological disease-causing variants and associated methylation signatures.
FULL-LENGTH RNA SEQUENCING OF ALZHEIMER’S
Long-read mRNA sequencing with the Iso-Seq method is used to sequence an Alzheimer’s…
EXPLORE
Did you know we have a more comprehensive library of articles, reports, papers, and videos related to human genomics?
UNCOVERING THE UNSEEN IN HUMAN GENOMIC RESEARCH SEQUENCING
Learn how these new applications will enable you to:
- More accurately identify human disease-causing tandem repeat changes than ever before with TRGT, a software solution for state-of-the-art tandem repeat analysis.
- More easily and cost-effectively obtain detailed sequences of just the genes you care about at scale, with the power of targeted HiFi sequencing and custom gene panels developed by Twist Bioscience.
UNLOCK YOUR NEXT GREAT DISCOVERY IN HUMAN GENOMICS
36,000 REASONS TO USE HIFI SEQUENCING
In 2019, scientists at the Mayo Clinic identified 36,794 dark regions in 6,054 gene bodies from pathways important to human health, development, and reproduction based on standard whole-genome short-read sequencing data.1 There are potentially thousands of important regions that can be missed without HiFi sequencing.
HIFI SEQUENCING REVEALS A MORE COMPLETE HUMAN REFERENCE GENOME
The complete, gapless human genome marks a new era of genomics where no region of the genome is beyond your reach. In 2021, PacBio HiFi technology was used by the Telomere-to- Telomere (T2T) Consortium to fill in those gaps.
ACHIEVE FEWER ERRORS DURING VARIANT DETECTION
PacBio HiFi technology was found to have the fewest total errors when compared to other technologies by the PrecisionFDA’s Truth Challenge V2: Calling variants from short and long reads in difficult-to-map regions.5
IDENTIFY INDELS WITH CONFIDENCE
A 2019 University of Washington study found that insertion-deletion mutations (indels) are routinely missed by short-read next generation sequencing (NGS) and long reads deliver 2.48 times as many structural variants as short reads alone, even “at their maximum capacity.” It is estimated that “at least 48% of deletions and 83% of insertions are routinely missed by the application of multiple short-read-calling algorithms.”2
SEE MORE STRUCTURAL VARIANTS
The 1000 Genomes Project sequenced 32 diverse reference genomes with PacBio technology, resulting in the de novo assembly of high-quality haplotype-resolved human genomes without parent-child trio data, or 64 assembled haplotypes from 32 genomes. Of the 107,590 structural variants identified, 68% were not discovered with short-read sequencing.4
OVERCOME PGX CHALLENGES
Interrogating CYP2D6 using short-read sequencing is challenging because of the degree of variation at the CYP2D6 locus and homology with its pseudogenes. Long-read sequencing can overcome these challenges.6

1. “Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight” Fryer et al. Genome Biol. 2019 May 20;20(1):97
2. “Genetic Variation, Comparative Genomics, and the Diagnosis of Disease.” Eichler EE. N Engl J Med. 2019;381(1):64-74
3. A complete human genome sequence is close: how scientists filled in the gaps https://www.nature.com/articles/d41586-021-01506-w
4. Ebert, Peter & Audano, Peter & Zhu, Qihui & Rodriguez-Martin, Bernardo & Porubsky, David & Bonder, Marc Jan & Sulovari, Arvis & Ebler, Jana & Zhou, Weichen & Mari, Rebecca & Yilmaz, Feyza & Zhao, Xuefang & Hsieh, PingHsun & Lee, Joyce & Kumar, Sushant & Lin, Jiadong & Rausch, Tobias & Chen, Yu & Ren, Jingwen & Eichler, Evan. (2021). Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science. 372. eabf7117. 10.1126/science.abf7117.
5. https://precision.fda.gov/challenges/10
6. Yang Y, Botton MR, Scott ER, Scott SA. Sequencing the CYP2D6 gene: from variant allele discovery to clinical pharmacogenetic testing. Pharmacogenomics. 2017 May;18(7):673-685.
“The SOLVE-RD team knows that long-read HiFi sequencing is
essential
for discovering the causal elements that have proven elusive with previous approaches.”
— Alexander Hoischen, PhD, Associate Professor for Genomic Technologies and Immuno-Genomics and a member of the SOLVE-RD team at Radboud University Medical Center
SHAPING THE FUTURE
CHILDREN’S MERCY KANSAS CITY FINDS INSIGHTS
Children with complex rare diseases are at the forefront of clinical research discoveries at Children’s Mercy Kansas City. See how scientists are moving beyond microarrays and exomes to enable improved care.
APPLICATIONS TO FUEL HUMAN GENOMICS RESEARCH
WHOLE GENOME SEQUENCING
Generate more complete and phased human genome assemblies of diverse populations to better understand the complexity of health and disease.
RNA SEQUENCING
Bring certainty to isoform detection with long-read sequencing to decipher how genomic variants drive the phenotypic differences between health and disease.
TARGETED SEQUENCING
Accurately detect and uncover more variant types even in hard-to-reach regions of the genome.
STRUCTURAL VARIANT DETECTION
Resolve variants at every scale to improve the power to link genetics to phenotypes of interest for novel discovery of genes and probable causative variants.
HUMAN GENOME MAPPING TO UNDERSTAND HUMAN HEALTH AND DIVERSITY
PAVE A PATH TO DIAGNOSTIC DISCOVERY
With 99.9% accuracy and long read lengths up to 25 kb, HiFi sequencing allows scientists to find possible causative pathogenic variants and identify novel disease-associated genes through:
Best-in-class variant calling for all variant types
HiFi sequencing includes solutions for single nucleotide variants (SNV), indels, copy number variants (CNVs), structural variants (SVs) and repeat expansions.
More complete, accurate, and phased assemblies of the human genome
Sequence regions previously inaccessible to other technologies — for accurate typing of genes including HLA and CYP2D6.
Uniform coverage
Achieve the coverage needed to access a more complete size spectrum of cancer mutations so that you can:
- Reveal patterns of structural variants
- Characterize genotypic differences between cohorts that respond differently to treatment by phasing distant SNPs or identifying structural variants
- Look beyond SNVs and robustly detect all structural variants to reveal novel insights not possible with short-read sequencing
IDENTIFY GENETIC DRIVERS OF DISEASE
To identify the genetic drivers of diseases, scientists must have access to a more complete genomic landscape, including regions previously deemed nearly impossible to sequence, and the tools to examine the full range of genomic variation.
The PacBio systems using Single Molecule, Real-Time (SMRT) sequencing provide exceptionally long reads, uniform coverage, and high consensus accuracy. This technology enables scientists to gain a more comprehensive understanding of the genetic basis of disease.
UNDERSTAND HOW TO HARNESS THE BENEFITS OF MEDICATION
Pharmacogenomics (PGx) impacts patient care by utilizing genomic information to assess an individual’s response to certain medications. PGx can be used to predict adverse drug reactions or decreased efficacy, leading to improved health outcomes and decreased costs. An estimated 99% of people have actionable variants in PGx genes,1,2 and studies have shown that up to 55% of adults2 are prescribed drugs for which there are PGx guidelines available. HiFi sequencing provides high-resolution insight into PGx loci, which include complex variants such as pseudogenes, tandem repeats, and CNVs.
2. Chanfreau-Coffinier C, Hull LE, Lynch JA, DuVall SL, Damrauer SM, Cunningham FE, Voight BF, Matheny ME, Oslin DW, Icardi MS, Tuteja S. Projected prevalence of actionable pharmacogenetic variants and level A drugs prescribed among US Veterans Health Administration pharmacy users. JAMA Netw Open. 2019 Jun 5;2(6):e195345. doi: 10.1001/jamanetworkopen.2019.5345. PMID: 31173123; PMCID: PMC6563578.
UNCOVER THE COMPLEXITIES OF IMMUNITY
From SNVs to structural variants, isoforms and somatic hypermutation, see the full range of variation that impacts the immune response in infectious disease, cancer, or autoimmunity.
The loci encoding the critical components of the vertebrate immune system are at once highly variable from person to person and hard to sequence. Only HiFi sequencing combines single-molecule accuracy with long read lengths, allowing you to:
- Generate fully phased haplotypes of immunologically important NGS dead zones including the KIR and IGH loci
- Determine HLA haplotypes unambiguously and without imputation
- See the full diversity of the adaptive immune response in action with full- length transcripts and isoforms in targeted, bulk, or single-cell experiments
FEATURED SEQUENCING SYSTEMS
Our portfolio of sequencers drive improvements in rare disease research, inspire personalized approaches to medicine, and enable the promise of genomics to better human health.