Nanobind extracted DNA is optimized for HiFi sequencing. Our kits provide you with an all-in-one extraction experience that can be used with a wide range of sample types. The PanDNA kit supports manual high-quality extraction from cultured cells, bacteria, whole blood, saliva, tissue, plant nuclei, and insect samples in 1 to 2.5 hours. The Nanobind HT CBB kit enables automation of DNA extraction for human and animal blood, mammalian cells, and bacteria. Our HT kit is compatible with up to four robotic platforms and 96 samples that can be extracted in 2.5 hours.
Nanobind PanDNA HMW DNA extraction protocols
How does it work?
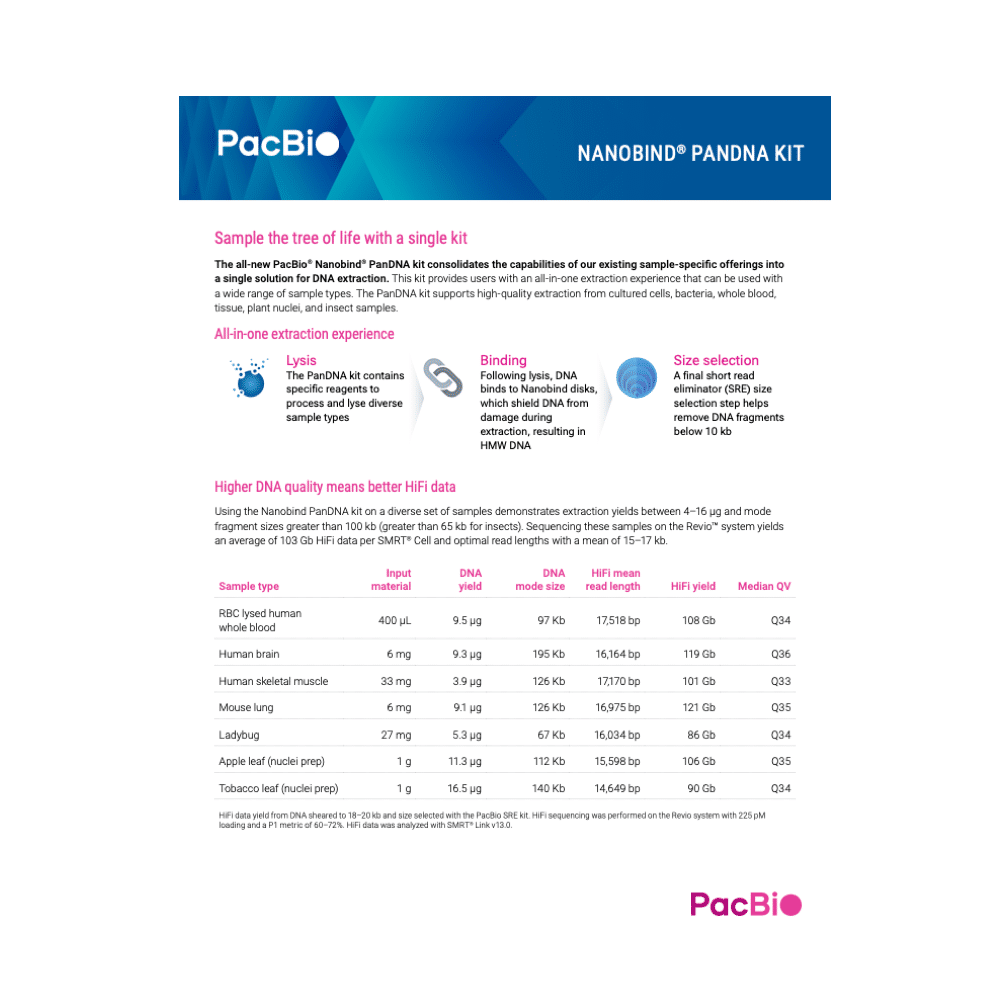
Nanobind kits contain specific reagents to process and lysate diverse sample types. Following lysis, the DNA binds to Nanobind disks which shield bound DNA from damage during extraction, resulting in HMW DNA. A final Short Read Eliminator (SRE) size selection step helps remove DNA fragments below 10 kb.
Brochure
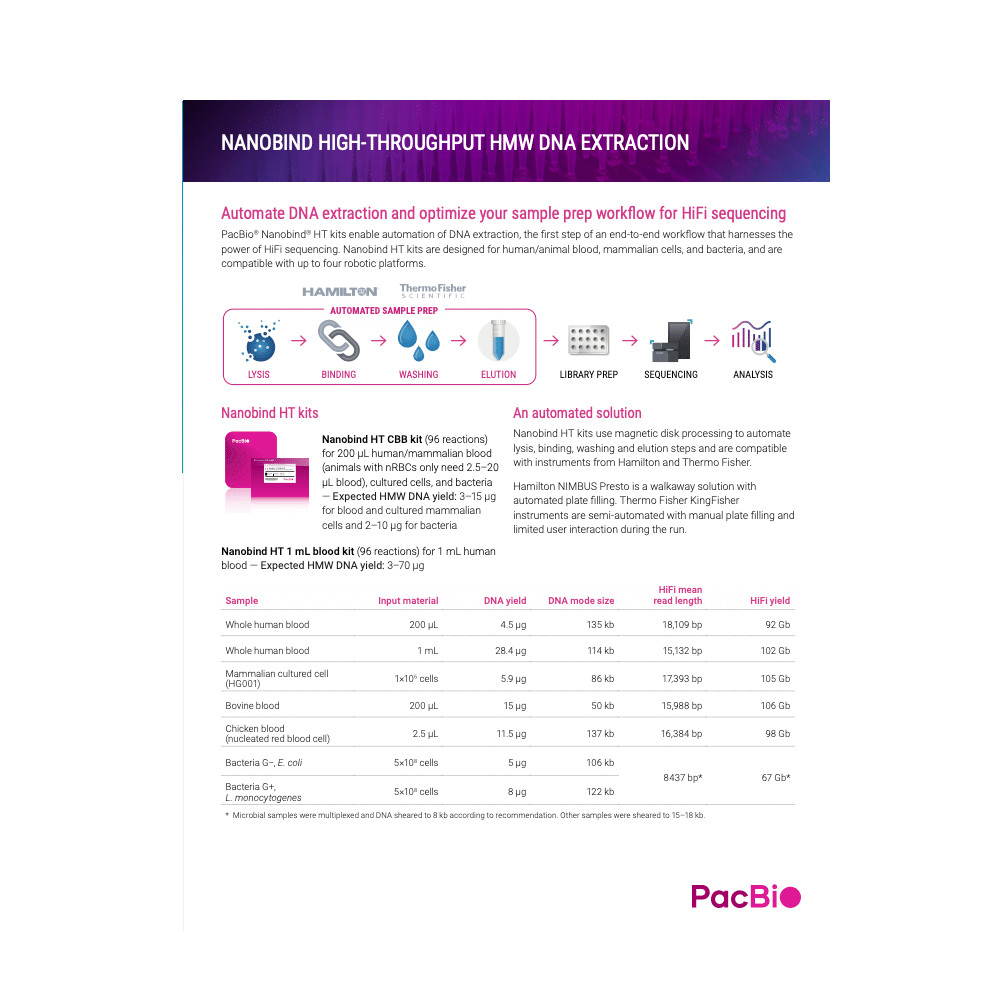
Automate DNA extraction and optimize your sample prep workflow for HiFi sequencing
PacBio Nanobind HT kits enable automation of DNA extraction, the first step of an end-to-end workflow that harnesses the power of HiFi sequencing. Nanobind HT kits are designed for whole human blood and mammalian cell samples and are compatible with four robotic platforms.
SHORT READ ELIMINATOR (SRE) KITS
Simple size selection for optimal HiFi result
SRE treatment following DNA extraction helps removing DNA molecules below 10 kB. It results in improved HiFi read length distribution and HiFi sequencing yield. The SRE workflow is fast, easy, and inexpensive using simple size selective precipitation. HMW DNA is precipitated by centrifugation and low molecular weight (LMW) DNA is washed away. Many samples can be process in parallel in 2 hours (10 min hands-on time). HT solution will be available soon.
Procedure + checklist
Removing short DNA fragments with the Short Read Eliminator (SRE) kit
This procedure describes the workflow for DNA size selection to remove molecules <10 kb using the SRE kit on HMW DNA before shearing and library preparation. The SRE kit is used for rapid size selection of unsheared HMW DNA samples. This method can significantly enhance mean read length by progressively depleting short DNA up to 10 kb.
PacBio compatible products
MAXIMIZE THROUGHPUT BY AUTOMATING LIBRARY PREP WITH PACBIO COMPATIBLE AUTOMATION PARTNERS
PacBio Compatible Kingfisher extraction scripts
NANOBIND KITS IN ACTION
NANOBIND
Nanobind PanDNA kit
The Nanobind PanDNA kit supports HMW DNA extraction from cells, bacteria, blood, tissue, plant nuclei, and insect.
Order kits online View brochure
Procedure + Checklists
Quick and easy reference guides to learn all about Nanobind kits and how they enable HMW DNA extraction.

Guides + Overviews
Nanobind guides provide recommendations for hard-to-sequence sample types.

END-TO-END WORKFLOWS FOR LONG-READ SEQUENCING AT SCALE
Extract the highest quality HMW DNA using Nanobind DNA extraction kits.
Enhance read lengths by depleting short DNA using SRE size selection kits.
Our experts offer end-to-end support from extraction through library prep and sequencing.