The paradigm shifts in human genetics research that are powered by PacBio sequencing have been described in numerous publications, and we have highlighted some of these previously, e.g. for the human pangenome reference, resolving even the most complex regions, and for advancing precision medicine. The same transformation is taking place for model organisms, generating much more complete and accurate resources for the scientific community that rely on them for their research. The latest example is a preprint entitled Resolution of structural variation in diverse mouse genomes reveals chromatin remodeling due to transposable elements by researchers from the University of Connecticut Health Center, The Jackson Laboratory and the University of Washington, applying PacBio sequencing to the mouse, and presenting data that are “an important step towards modernizing mouse genetics.”
The authors highlight that diverse inbred mouse strains are among the foremost models for biomedical research, yet genome characterization of many strains has been fundamentally lacking in comparison to human genomics research. In particular, the cataloging of structural variants (SVs) is incomplete, limiting the discovery of potentially causative alleles for phenotypic variation across individuals, which is attributed to the shortcomings of short-read sequencing: “Despite recent efforts to construct strain-specific reference genomes, much of the genomic landscape that is variable between these strains remains incomplete due to the technological limitations of short-read whole genome sequencing.”
In the new study, the researchers utilized PacBio sequencing to resolve genome-wide SVs in 20 genetically distinct inbred mice. Generating de novo assemblies, they have created “the most contiguous genome assemblies of diverse mouse genomes produced to date” – 534x more contiguous, 143x fewer contigs, and 228 Mb of additional sequence on average compared to short-read assemblies!
Leveraging these high-quality assemblies for variant calling, they report over 400,000 site-specific SVs that affect 13% (356 Mb) of the current mouse reference assembly, “including over 500 previously unannotated variants which alter coding sequences.” In contrast, “short-read SV calling was only able to detect 46% of deletions, 14% of insertions, and 39% of inversions discovered by long-read sequencing.”. In comparison to the previous resource, over 215,000 SVs are described “which are absent from public mouse SV catalogs.”
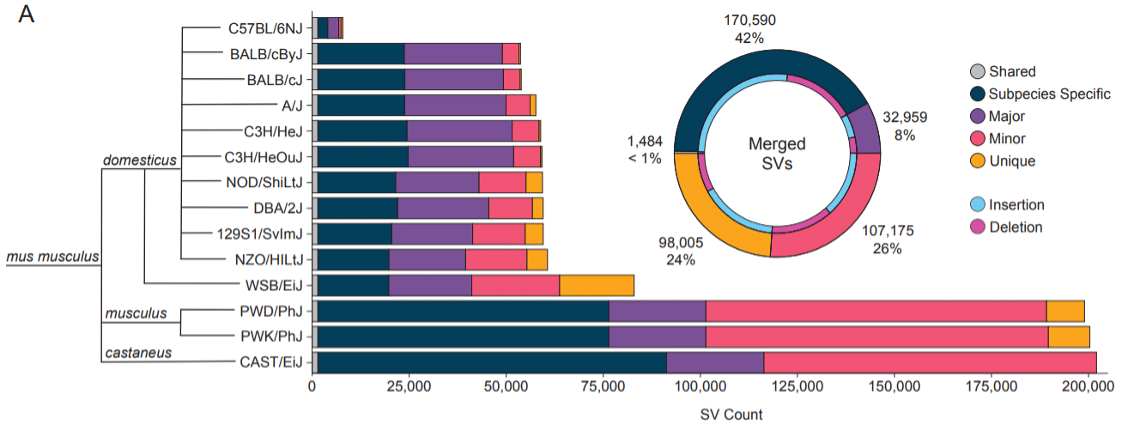
Figure 1A from Ferraj et al.: “Discovery of Structural Variants in Diverse Mouse Genomes (A) Total number of structural variants discovered in each mouse genome when compared to the mus musculus (GRCm39, C57BL/6J background) reference genome. Variants are grouped by their frequency within the cohort: shared (present in all strains) Major (≥ 50% of the cohort), Minor (< 50%), exclusive to one strain (Unique) or exclusive to one subspecies (Subspecies specific). We merged variants into a non-redundant callset (donut plot), shown along with the proportion of insertion and deletion calls (blue and purple).”
The researchers conclude that “the full potential of mouse genetic reference populations cannot be fully realized until SVs are entirely sequence resolved. We have made important steps in rectifying this deficit.”
Please connect with us if you are interested in overcoming similar deficits in the genetic characterization of your favorite (model or non-model) organisms.