Antibodies are the work horses of the adaptive immune system, identifying and neutralizing foreign objects like bacteria and viruses, and thereby are central to the science of immunology. A new preprint by Dr. Melissa Smith & colleagues from the University of Louisville, entitled FLAIRR-seq: A novel method for single molecule resolution of near full-length immunoglobulin heavy chain repertoires, describes a new gold standard to characterize expressed antibody repertoires.
The authors highlight that previous methods for Adaptive Immune Receptor Repertoire Sequencing (AIRR-seq) have limited resolution: “All commonly used AIRR-seq methods are … technically limited by the length restrictions (≤600nt) of short-read next generation sequencing. As a result, no current AIRR-seq strategy resolves the complete heavy chain transcript.” Taking advantage of the combined length and accuracy of HiFi reads, the researchers extended the reach of sequencing to include most of the constant region of immunoglobulin (IG) heavy chain transcripts. It is based on “amplification of near full-length IgG and IgM heavy chain transcripts, paired with SMRT sequencing, resulting in highly accurate (mean read accuracy ~Q60, 99.9999%), near full-length Ab sequences”. This novel, near Full-Length AIRR-seq (FLAIRR-seq) method “provides, for the first time, simultaneous, single-molecule characterization of IGHV, IGHD, IGHJ, and IGHC region genes and alleles, allele-resolved subisotype definition, and high-resolution identification of class-switch recombination within a clonal lineage.”
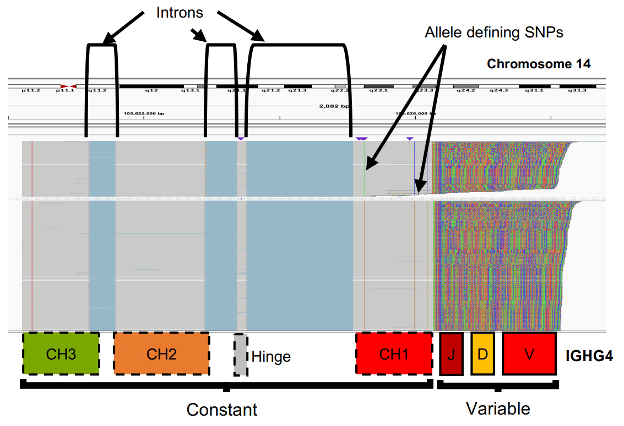
After benchmarking the method against matched datasets generated by previous methods, demonstrating “robust, unbiased FLAIRR-seq performance using RNA samples derived from peripheral blood mononuclear cells, purified B cells, and whole blood”, and “additionally resolving novel IG heavy chain constant (IGHC) gene features.”, the researchers applied FLAIRR-seq to 10 individuals, resulting in the identification of 32 unique IGHC alleles. Remarkably, 28 of these (87%!) were previously uncharacterized. Such a high discovery rate of “novel or extended alleles underscores the extensive polymorphism in this region and reflects the paucity of information regarding this locus in existing immunogenomics databases.” In addition, the researchers investigated the utility of FLAIRR-seq in clinical samples, particularly to observe changes in immune repertoires over time, and including the resolution of Ab repertoire changes in an individual hospitalized over several days for severe COVID-19 disease.
Having demonstrated the capabilities of FLAIRR-seq to characterize IG gene diversity for the most comprehensive view of bulk-expressed Ab repertoires to date, the authors conclude by stressing that “it is critical to account for all variability within the Ab repertoire for the most comprehensive understanding of repertoire dynamics and the myriad factors impacting Ab effector function”, including the potential for improved predictive markers for disease (including cancer), disease progression and the transition of acute to chronic disease states.
Interested in adding the flair of unparalleled sequencing accuracy to your research, in immunology or any other field? Please connect with us for help in designing your next research project.