
In the rapidly advancing field of genomics, choosing the right long-read sequencing technology is key to meeting your experimental goals smoothly and successfully. To make an informed decision, it’s important to understand how different long-read data types, namely HiFi reads and nanopore reads, stack up against each other based on your project’s specific needs—like accuracy, application fit, cost, resources, and data accuracy. Here are a few examples of how these choices play out in real research scenarios:
- Human genomic studies: HiFi sequencing is your best bet if your project needs high accuracy to identify genetic variants and mutations. HiFi sequencing combines long read lengths with exceptional accuracy, even in the challenging “dark” regions of the genome, such as GC-rich or repetitive areas. HiFi sequencing can accurately call and phase both small and large variants, providing crucial haplotype information essential for disease research.
- Rapid pathogen identification: During disease outbreaks, speed is everything. Nanopore sequencers offer a rapid setup and real-time data generation, making it possible to quickly identify pathogens. This capability makes it a valuable tool for scientists and healthcare professionals managing infectious disease outbreaks.
What is long-read sequencing and why choose it over short-read solutions?
Long-read sequencing produces genomic data by generating individual reads that are thousands of nucleotides or more in length. These reads typically come from “native” DNA or RNA from a biological sample, preserving any base modifications present. On the other hand, most short-read sequencing technologies use fragments that are 50 to 300 bases long and rely on enzymatic amplification to generate sufficiently large clonal populations, which introduces bias and loses information about base modifications.
In the past decade, there have been efforts, such as Illumina’s Complete Long Reads, to create longer sequences from short sequence reads. We’ve discussed this in our posts “True long reads vs. synthetic long reads” and “Getting the right answer”, highlighting that Illumina Infinity/CLR reads contain many errors, leading to inaccurate results for researchers. Native long HiFi reads from PacBio are exceptional in providing comprehensive, accurate, and phased variant information, along with 5mC methylation data—which is not possible with Infinity/CLR due to amplification. Synthetic long reads consistently fall short compared to PacBio HiFi reads.
Comparing PacBio HiFi sequencing and Oxford Nanopore Technologies (ONT) and other Nanopore sequencing
How does nanopore sequencing work?
Nanopore sequencing technology, popularized by Oxford Nanopore Technologies, involves passing a single strand of DNA or RNA through a protein nanopore embedded in a membrane. A voltage applied across this membrane causes ions to flow through the pore, creating an electric current. The electrical current changes based on the piece of DNA or RNA passing through the tiny pore, with numerous nucleotides affecting the signal at the same time.1,5,6 To determine the DNA sequence from these signals, scientists match the changes in the current to known patterns of short DNA pieces. Because the current doesn’t show the identity of individual nucleotides, nanopore sequencing can sometimes make mistakes in calling indels within repetitive regions, where it can lose track of the sequence.
Nanopore sequencing can also be used to detect modified bases, since these modifications alter a nucleotide’s ability to influence current flow. This presents both an opportunity and a challenge. On the one hand, it provides an opportunity to detect a flexible set of possible base modifications. On the other hand, modified bases expand the set of possible answers to the question “what sequence explains my current signal?” making basecalling more difficult. Users often need to choose a basecalling model that balances sensitivity to likely modifications with overall accuracy and speed.

| Nanopore advantages | Nanopore disadvantages |
|---|---|
|
Ultra-long reads: Can produce reads that are sometimes over hundreds of thousands of bases long or even exceed a megabase. Portable sequencers: Instruments like the Oxford Nanopore MinION are portable and affordable, making them suitable for small-scale analyses. Versatility: Can sequence native DNA and RNA, including detecting RNA modifications, without the need for amplification. |
Lower accuracy and systematic errors: Lower raw read accuracy and systematic errors in low complexity sequence regions, leading to higher coverage requirements and persistent indel errors2,3,4. File storage and data processing costs: Large file sizes (1,300 GB) make storage expensive, and base calling can take days per genome, requiring costly GPU servers. |
How does HiFi sequencing work?
HiFi sequencing is unique because it is very accurate, with a 99.9% accuracy, comparable to other top methods like short reads and Sanger sequencing. Developed by PacBio, HiFi sequencing uses fluorescent light signals to identify DNA bases and modified bases without bisulfite treatment. As a polymerase enzyme adds new nucleotide bases to a newly replicated strand, it emits tiny flashes of light. This process occurs inside small wells on a special microchip called a SMRT Cell, which contains millions of these tiny wells.
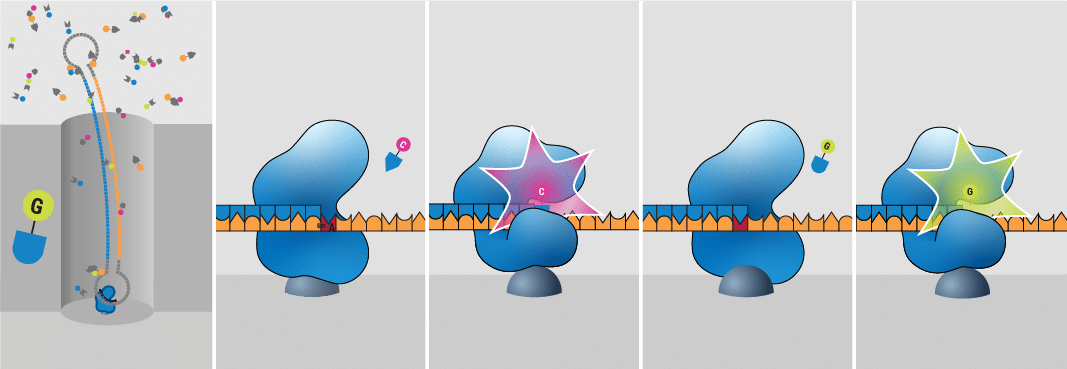
| HiFi advantages | HiFi disadvantages |
|---|---|
|
High accuracy: HiFi sequencing typically exceeds 99.9% accuracy, making it suitable for applications requiring high-quality base calling. Long reads: Provides 15,000 to 20,000 bases or more in read lengths to span large genomic regions and repeats. Comprehensive insights: Delivers high-quality reads of sequence and methylation status, including in regions not accessible to short-read technologies. |
Higher system cost: Systems are larger and more expensive than nanopore systems, though sequencing cost per genome is often more affordable, helped by lower coverage requirements. |
Choosing the appropriate long-read solution for your project
When choosing a long-read technology platform, consider the following:
- Accuracy and read length: Different technologies offer varying levels of accuracy and read lengths, which impact the quality of sequencing results.
- Application suitability: Some platforms are better suited for specific applications, such as detecting structural variants, RNA sequencing, or de novo genome assembly.
- Cost and resources: Budget constraints and available resources may influence your choice, as some platforms are more cost-effective or resource-intensive than others.
- Data analysis: The chosen technology influences the bioinformatics tools and pipelines required for data processing and analysis.
Table showing comparison of long read technologies
| PacBio HiFi sequencing | ONT Nanopore sequencing | |||
|---|---|---|---|---|
| Input | DNA, cDNA | DNA, RNA | ||
| Read length | 500 to 20 kb | 20 to >4 Mb | ||
| Read accuracy | Q33 (99.95%) |
~Q20 2,3,4 |
||
| Typical run time | 24 hours | 72 hours | ||
| Typical yield per cell | 90 Gb (Revio) |
50-100 Gb 7 |
||
| Base calling | Yes, on-instrument ($0) | Off-instrument, often requires additional costly GPU server | ||
| Variant calling - SNV | Yes | Yes | ||
| Variant calling - Indels | Yes | No | ||
| Variant calling - SVs | Yes | Yes | ||
| Detectable DNA modifications | 5mC on-instrument calling ($0) | 5mC, 5hmC, and 6mA. Off-instrument calling, often requires additional costly GPU server | ||
| Platforms | Revio, Sequel, and Sequel IIe | PromethION, MinION, GridION, and Flongle | ||
| Typical output file size (type) | 55 GB (BAM) | 1,300 GB (fast5/pod5) | ||
| Storage cost per month* | $1.30 USD | $30.00 USD | ||
| Table footnote: * AWS S3 Standard cost per month is calculated based on USD $0.023 per GB storage pricing. Oxford Nanopore Technologies information gathered from https://nanoporetech.com/platform | ||||
The long and short of it
Both PacBio HiFi sequencing and ONT nanopore sequencing have their strengths, and the choice depends on the specific needs of your project, such as read length, accuracy, cost, and application. For impactful science, HiFi sequencing is often the best fit due to its high accuracy and comprehensive insights. Beyond technical benefits, PacBio has a global and world-class service and support team that provides personalized assistance throughout the HiFi sequencing process, supporting scientists every step of the way.
Ready to get serious about long-read sequencing?
Learn more about PacBio HiFi sequencing:
See if HiFi sequencing can be applied to your applications
- Human genomics
- Microbial genomics
- Plant and animal genomics
- Cancer genomics
- Cell and gene therapy research
Connect with a PacBio scientist
References
- Deamer, D., Akeson, M. & Branton, D. Three decades of nanopore sequencing. Nat Biotechnol 34, 518–524 (2016). https://doi.org/10.1038/nbt.3423
- Clinical application of Complete Long Read genome sequencing identifies a 16kb intragenic duplication in EHMT1 in a patient with suspected Kleefstra syndrome. John E. Gorzynski, Shruti Marwaha, Chloe Reuter, Tanner D. Jensen, Alexis Ferrasse, Archana Natarajan Raja, Liliana Fernandez, Elijah Kravets, Jennefer Carter, Devon Bonner, Shirley Sutton, Undiagnosed Diseases Network, Maura Ruzhnikov, Louanne Hudgins, Paul G Fisher, Jonathan A. Bernstein, Matthew T. Wheeler, Euan A. Ashley. medRxiv 2024.03.28.24304304; doi: https://doi.org/10.1101/2024.03.28.24304304
- Harvey, W. T., Ebert, P., Ebler, J., Audano, P. A., Munson, K. M., Hoekzema, K., Porubsky, D., Beck, C. R., Marschall, T., Garimella, K., & Eichler, E. E. (2023). Whole-genome long-read sequencing downsampling and its effect on variant-calling precision and recall. Genome Research, 33(12), 2029–2040. doi:10.1101/gr.278070.123
- Mahmoud, M., Huang, Y., Garimella, K. et al. Utility of long-read sequencing for All of Us. Nat Commun 15, 837 (2024). https://doi.org/10.1038/s41467-024-44804-3
- Rang, F.J., Kloosterman, W.P. & de Ridder, J. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol 19, 90 (2018). https://doi.org/10.1186/s13059-018-1462-9
- Stoddart, D., Maglia, G., Mikhailova, E., Heron, A. and Bayley, H. (2010), Multiple Base-Recognition Sites in a Biological Nanopore: Two Heads are Better than One†. Angewandte Chemie International Edition, 49: 556-559. https://doi.org/10.1002/anie.200905483
- Sigurpalsdottir, B.D., Stefansson, O.A., Holley, G. et al. A comparison of methods for detecting DNA methylation from long-read sequencing of human genomes. Genome Biol 25, 69 (2024). https://doi.org/10.1186/s13059-024-03207-9