As the fastest growing neurological disease in the world, Parkinson’s Disease (PD) is prompting communities to find ever more creative ways to cope with the devastating toll of this disease. Therapeutic programs using dance, boxing, and even ping pong have been introduced to help combat the strength and motor coordination symptoms that accompany PD. These innovative initiatives allow patients to benefit from exercise therapy and, all-importantly, invaluable human connection.
As a testament to the power of community, what began as a personal hobby for Nenad Bach, the founding member of Ping Pong Parkinson, has grown into a supportive community. When asked about the value of community he stated, “We haven’t conquered Parkinson’s, but we conquered the fear of Parkinson’s.”
Scientific efforts towards conquering Parkinson’s
Right alongside the communities in this fight are scientists working toward conquering Parkinson’s itself. Promising therapies like neuron transplants, stem-cell therapy, and immune-based treatments are currently the focus of ongoing research.
New frontiers in Parkinson’s treatment: Antisense Oglionucleotides (ASOs)
Among the newest frontiers to promising Parkinson’s treatments is the development of antisense oligonucleotides (ASOs). ASOs are small, synthetic molecules that bind to mRNA sequences to alter protein expression. In the case of Parkinson’s Disease, ASOs most often target the protein alpha-synuclein (aSyn), a key protein that aggregates in the brain of Parkinson’s patients, causing a decline in motor and cognitive function. ASOs have shown early success in animal models and are currently in use through a phase I clinical trial.
Importance of mRNA isoforms in ASO research
Because ASOs bind to mRNA transcripts, a complete and accurate picture of the different splice variants of the same gene (transcript isoforms), is vital for the research and development of this promising therapy. Currently the PacBio Iso-Seq method is one of the best methods to offer the accuracy needed to deliver this comprehensive view of the transcriptome.
In this blog, we highlight how a team of researchers in UK, in collaboration with the Aligning Science Across Parkinson’s (ASAP) Collaborative Research Network recently used the PacBio Iso-Seq method to map the transcriptional diversity of Parkinson’s and develop ASOs with the ability to reverse PD-associated pathology in an established cell model.
Focus on SNCA gene coding for alpha-synuclein protein
In this study, a team led by researchers across University College London, University of Cambridge, the Francis Crick Institute, and the ASAP Network performed targeted sequencing using the Iso-Seq method to characterize the transcripts of the SNCA gene, which encodes for the key aSyn protein. They identified 42 unique SNCA transcripts in PD neurons, of which only 2 had been previously annotated, revealing that over 95% of the transcripts they found are novel.
Next, the researchers zeroed in on the sections of the SNCA gene that specifically code for the aSyn protein, rather than purely regulatory sections that control how genes are expressed. Ten percent of these regions were also previously unidentified, and the researchers were able to use calculations to predict how these regions would translate to aSyn proteins with specific characteristics that may affect PD pathology.
Role of regulatory elements in the SNCA gene related to Parkinson’s
However, beyond just looking at the coding region of SNCA, it’s important to not overlook the regulatory elements of the SNCA gene. Earlier evidence has found that these ‘untranslated regions,’ or UTRs, also play central roles in the development of Parkinson’s. Remarkably, the study that validated these findings found that 75% of SNCA gene expression was found to originate from novel ‘alternative’ UTRs. These UTRs have varied sequence lengths, also acting to regulate the expression of the SNCA gene.
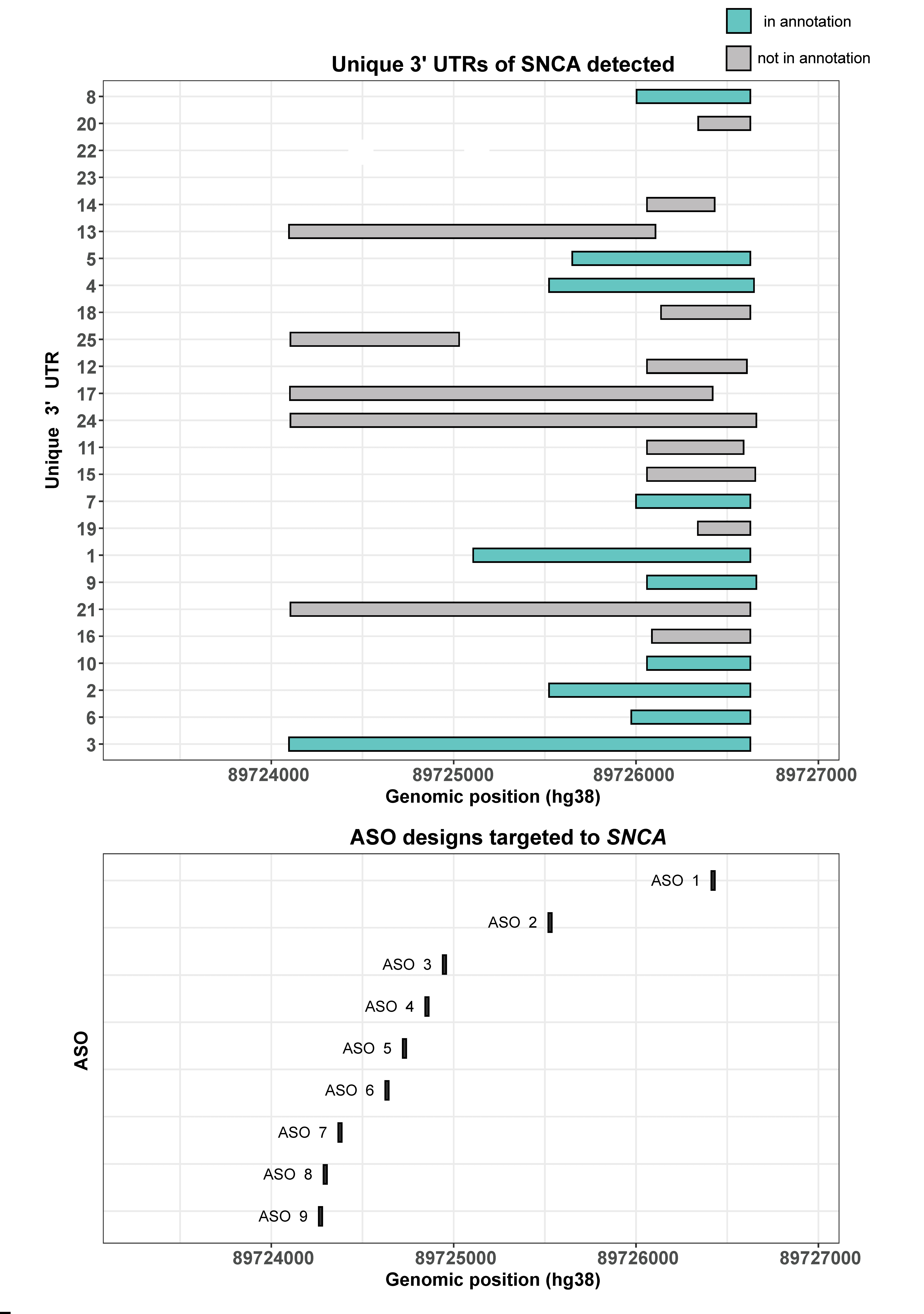
Figure 1, Evans, Gustavsson, et al., 2024. SNCA transcript structures detected in iPSC-derived mDA neurons and the targeting location of the 9 ASOs designed.
By precisely characterizing the 3’ UTR, the UK team was able to create several ASO candidates that reduced SNCA gene expression. Impressively, this effect lasted, with the strongest impact at 10 days for ASO-1-MOE. It significantly reduced aSyn protein aggregation and reversed PD-associated cellular pathology, including mitochondrial dysfunction and cell death.

Figure 2N, Evans, Gustavsson, et al., 2024. Live-cell images depicting dead cells in SNCAx3 mDA neurons with and without SNCA-targeting ASO treatment using the fluorescent dye SYTOX green.
The PacBio difference in sequencing accuracy
These remarkable results were possible only because of the outstanding sequencing accuracy provided by PacBio HiFi long-read sequencing technology. The researchers pointed out that, had they used a less precise ASO sequence targeting the typical 3’ UTR rather than the alternative sequence revealed by the PacBio Iso-Seq method, it would “only target a small proportion of total SNCA expression.”
This study clearly demonstrates how the high accuracy of PacBio technology can significantly impact the development of potential new therapies. The authors encourage a more comprehensive exploration of the PD isoform landscape as a way to bolster potential treatments like ASOs, noting that “a more complete understanding of disease-relevant transcript structures may ultimately enable the generation of ASOs that specifically target disease-associated structures.”
Broader applications of HiFi sequencing
But the story doesn’t stop there. This study on Parkinson’s Disease is just one example of how PacBio sequencing technology is making a significant impact on neurodegenerative disease research. Earlier this month, the Australian bioinformatics company GenieUs Genomics announced the integration of its DiGAP tool with PacBio HiFi sequencing on the Revio system for ALS research. This Deep integrated Genomic Analysis Platform uses machine learning to classify HiFi long-read sequencing data from ALS patients into distinct genetic profiles, enabling the development of personalized treatments. In collaboration with Duke and Temple Universities, this initiative aims to provide personalized experimental therapies to 50 ALS patients based on their unique DiGAP classifications.
Experts agree that the accuracy of PacBio HiFi technology is fundamental to advancing these types of genetically nuanced and personalized approaches.
Conquering fear and disease with science and community
We are inspired by these findings and confident that through harnessing highly-accurate sequencing technology and fostering a community of research and understanding, we are moving closer to conquering not only the fear of neurodegenerative diseases but the diseases themselves.