Transform your clinical lab with innovative precision
and unmatched efficiency
Consolidate and streamline multiple genetic tests on a single platform. PacBio HiFi sequencing supports the development of genetic tests by providing unbiased coverage, minimizing the need for re-testing and supplemental assays when new variants are discovered, and allowing more focus on operational efficiency at scale.
HIGH-QUALITY, HIGH-EFFICIENCY, HIFI SEQUENCING ENABLES:
- Consolidation of multiple assays
- Scalable, automation friendly library prep solutions
- Methylation detection without any additional workflows
- Complete, accurate, haplotype-phased variant calling in clinically-relevant regions of the genome
- Discovery power for applications like pharmacogenomics (PGx), HLA, carrier screening and hereditary cancer screening
Targeted sequencing
Comprehensively sequence clinically-relevant genes of interest cost-effectively at scale.
PureTarget
Comprehensively genotype targeted repeat loci at scale to power neurological and carrier screening research.
Clinical research
Get detailed information into complex genetic regions by detecting structural variants and repeat expansions to accelerate personalized medicine in clinical research.
Population Genomics
Utilize scalable long-read sequencing to explore genomic variation, driving precision medicine initiatives in diverse populations.
Targeted sequencing for complex genes
With targeted sequencing, you only sequence the genomic regions you care about. HiFi targeted sequencing offers flexible and scalable sequencing of specific genomic regions, with hybrid capture and amplicon workflows available for your customized needs.
HiFi reads provide accurate haplotype resolution and comprehensive detection of all variant types, while also providing access to difficult-to-sequence regions like those with high homology, GC-rich and repetitive regions. It can also resolve complex genes and pharmacogenes such as SMN1/2, FMR1, GBA, HBA, LPA, PMS2, and CYP2D6.
Blog
Get more for less — Single-gene testing with HiFi sequencing
For cost-effective assay consolidation, with all the benefits of HiFi long-read sequencing, amplicon workflows can be useful for high-volume testing of clinically-relevant genes.
Spotlight
Closing the gap
Authors describe how HiFi targeted enrichment-based sequencing offers a cost-efficient and scalable approach to assess 389 medically-relevant yet challenging “dark” genes. Mahmoud M, et al. (2024) Closing the gap: solving medically-relevant genes at scale
Application note
Enabling thalassemia genotyping at scale
Screening for thalassemias involves genotyping of HBA1 and HBA2 (~97% homologous) and HBB, which can be difficult to resolve because of indels, SVs, CNVs, and the need for phasing, as well as rare or novel variants that are present in different ethnicities. Learn more about how amplicon-based HiFi sequencing provides an end-to-end workflow for enabling high-volume thalassemia tests.
Poster
Enabling scale with automated library prep solutions
Learn more about automated protocols for DNA shearing and size selection, which remove library prep bottlenecks, improve throughput, and dramatically lower cost.
HiFi Sequencing for Pharmacogenomics
HiFi sequencing, provides high-resolution insight into PGx loci, which include complex variants such as pseudogenes, tandem repeats, and CNVs. Compared to other technologies, long-read HiFi sequencing allows for accurate detection of structural and complex variants, direct haplotype phasing, and coverage of rare variants. With read lengths of up to 25 kb and 99.9% accuracy, HiFi sequencing enables PGx discovery and translational research to help drive personalized medicine and potentially improve patient care.
References:
- McInnes G, Lavertu A, Sangkuhl K, Klein TE, Whirl-Carrillo M, Altman RB. Pharmacogenetics at Scale: An Analysis of the UK Biobank. Clin Pharmacol Ther. 2021 Jun;109(6):1528-1537. doi: 10.1002/cpt.2122. Epub 2020 Dec 17. PMID: 33237584; PMCID: PMC8144239.
- Chanfreau-Coffinier C, Hull LE, Lynch JA, DuVall SL, Damrauer SM, Cunningham FE, Voight BF, Matheny ME, Oslin DW, Icardi MS, Tuteja S. Projected Prevalence of Actionable Pharmacogenetic Variants and Level A Drugs Prescribed Among US Veterans Health Administration Pharmacy Users. JAMA Netw Open. 2019 Jun 5;2(6):e195345. doi: 10.1001/jamanetworkopen.2019.5345. PMID: 31173123; PMCID: PMC6563578.
Spotlight
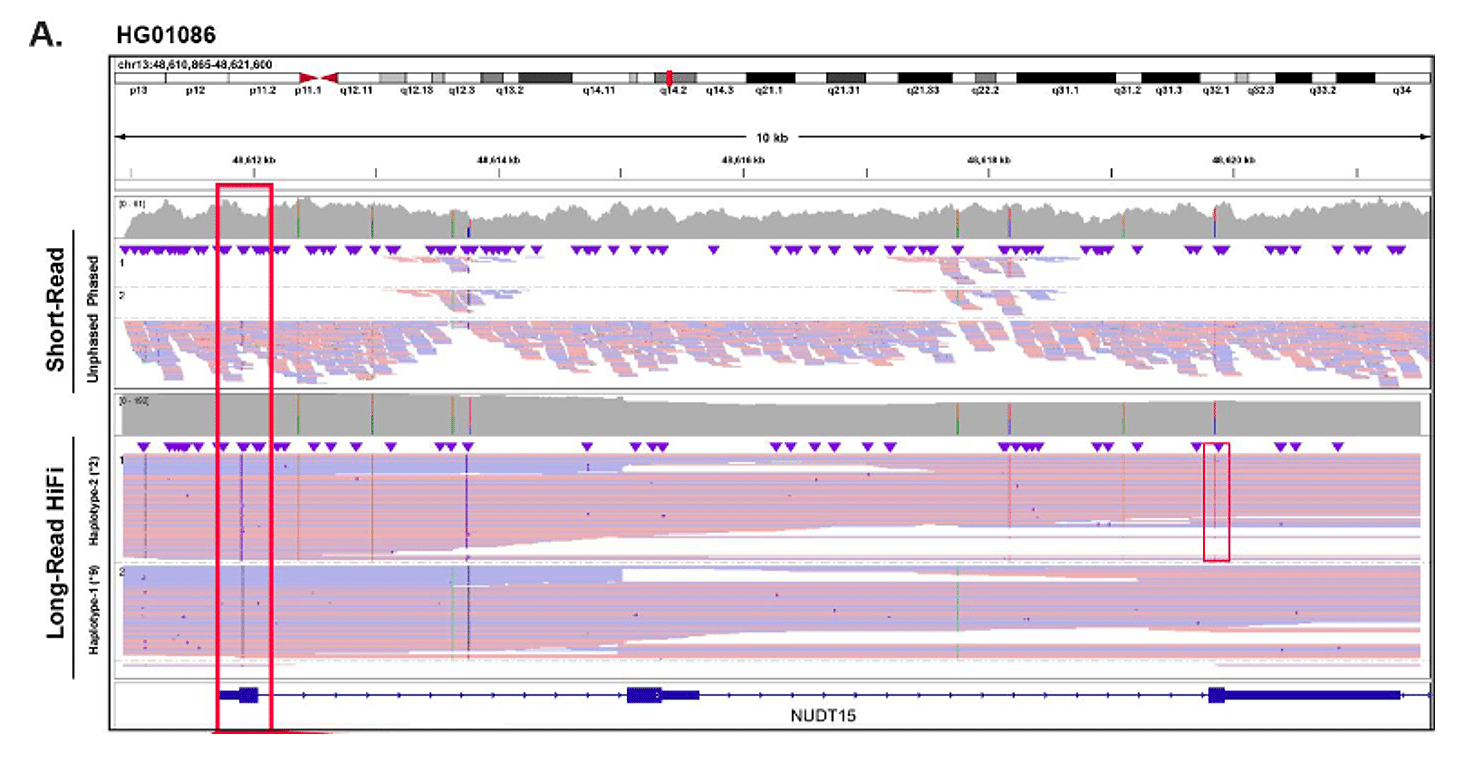
High resolution phasing of star alleles improves prediction to drug response
HiFi amplicon sequencing was used in this study to sequence and phase NUDT15 variants for accurate star (*) allele diplotyping, which is associated with response to thiopurine drugs. This included a sample that was previously mischaracterized by short-read sequencing due to phasing ambiguity, which resulted in an incorrect predicted drug response phenotype. Additionally, HiFi sequencing enabled novel star (*) allele discovery (NUDT15*1.007, *20) as well.
HiFi sequencing for pharmacogenomics in action
Application note
HiFi amplicon sequencing for pharmacogenetics: CYP2D6
With PacBio HiFi sequencing, CYP2D6 variation can be captured using highly accurate long reads on a single platform. Amplicon-based HiFi sequencing fully and directly resolves and phases complex loci like CYP2D6 without assembly or inference, allowing for ancestry agnostic haplotype assignment. In this CYP2D6 application note, we propose a streamlined PCR amplicon workflow that characterizes full-length CYP2D6 alleles with PacBio HiFi sequencing.

Highly scalable pharmacogenomic panel testing with hybrid capture and long-read sequencing
With long-read PacBio HiFi sequencing and Twist Bioscience hybrid capture technology, we describe a pre-designed PGx panel that is comprehensive and cost-efficient, allowing for scalable application to precision medicine research programs.

Enabling research in pharmacogenomics
With robust coverage of difficult-to-sequence and difficult-to map regions, combined with high accuracy variant calling and unambiguous haplotype resolution through direct phasing, HiFi sequencing is a powerful tool in PGx.

From sample to star alleles
We demonstrate the use of the Twist Alliance Long-Read PGx panel for use with PacBio HiFi sequencing systems, with an analysis pipeline that includes primary analysis (on-instrument), bioinformatics tools for secondary and tertiary analysis, star allele calling with Pangu and PharmCAT, and interpretation and reporting with PharmCAT and custom EHR clinical decision support (CDS) (e.g. Epic Best Practice Alerts).
Explore
Did you know we have a comprehensive library of reports, papers, and videos related to genetic testing?
Access the full spectrum of polymorphisms across the HLA system
Highly accurate long reads — HiFi reads — with single-molecule resolution make Single Molecule, Real-Time (SMRT) sequencing technology ideal for generating unambiguous, phase-resolved imputation-free HLA typing, so that you can:
- Span complete HLA class I genes and long amplicons of HLA class II allowing for four field HLA genotyping
- Fully phase polymorphisms with allele specificity across SNP-poor regions of HLA genes
- Achieve unambiguous allele-level segregation without imputation
- Detect variants in regulatory regions within 5’ UTRs, introns, and 3’ UTRs
- Fully characterize minor variants in polyclonal samples, such as cancer or transcripts
- Obtain direct evidence for new HLA alleles through de novo, reference-free consensus generation
Blog
SMRT Sequencing Provides Higher-Resolution HLA Typing Associated with Improved Patient Survival Rates
In a study just published in the Journal of Biology of Blood and Marrow Transplantation, scientists at the Anthony Nolan Research Institute demonstrated that ultra-high-resolution HLA typing performed with SMRT sequencing identified stronger matches associated with improved survival rates among patients who received hematopoietic cell transplants.
Spotlight
Creating the gold standard for HLA reference alleles
The IPD-IMGT/HLA database has provided the community with a repository of observed HLA alleles and sequences for over 20 years. This resource is used to assign official allele designations, which have implications in clinical research and genetic testing of HLA loci. Since “long read sequencing is able to generate complete fully phased sequences for an entire HLA gene” (Barker, et al. Nucleic Acid Res (2023)), PacBio has been instrumental in the development and expansion of this critical resource.