 Whether it’s enhancing marker development, facilitating the production of more nutritious food, protecting livestock health, or increasing crop yield, the biological insights gained from comprehensive HiFi sequencing are immeasurable for the agricultural industry.
Whether it’s enhancing marker development, facilitating the production of more nutritious food, protecting livestock health, or increasing crop yield, the biological insights gained from comprehensive HiFi sequencing are immeasurable for the agricultural industry.
Following AGBT AG in the first of a three-part series, we’ll explore how PacBio provides cutting-edge sequencing solutions that enable agricultural scientists to sustainably meet a growing demand to feed the world.
A new breed of sequencing tools
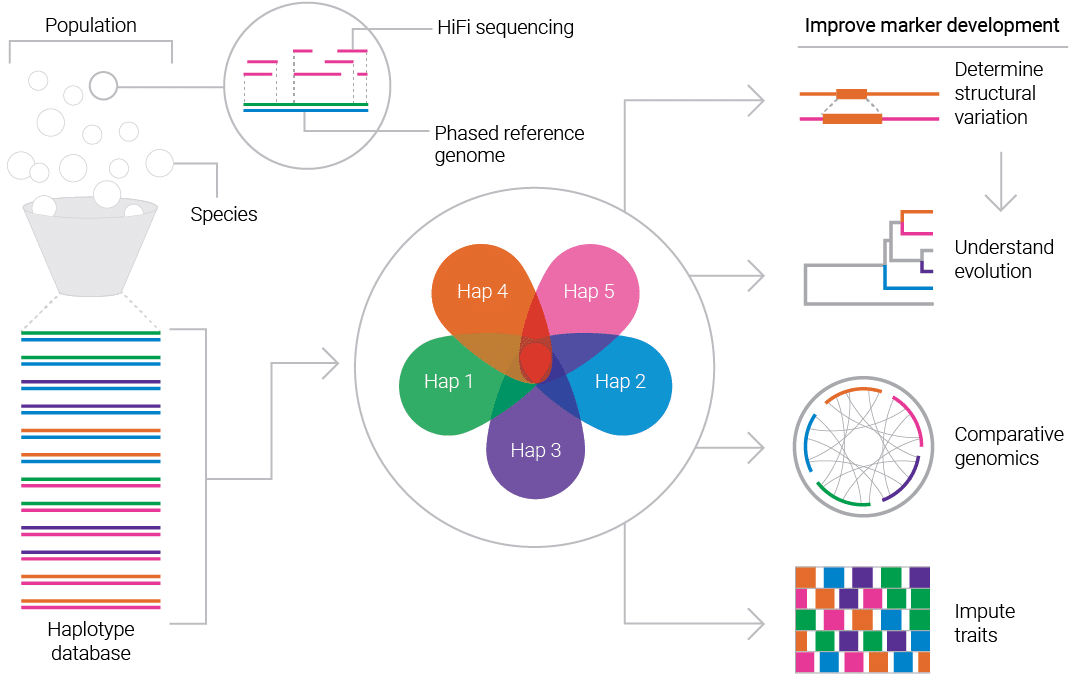
Whole genome sequencing has provided breeders with data-driven strategies for marker-assisted selections by identifying desirable traits. However, plant and animal biology is complex and these traits are often too complex to use single SNPs for imputation. They require understanding at a complete haplotype scale.
Because HiFi sequencing is single-molecule in nature with highly accurate reads of lengths that span many kilobases, it is an ideal tool to quickly assemble phased, reference-quality genomes for even the most complex plant and animal genomes.
Thanks to HiFi sequencing, scientists are able to:
- Impute desirable traits to SNPs, structural variants, and complex genotypes
- Capture genomic variants on a genome-wide scale for outbreds, inbreds, and populations
- Build reference-quality, haplotype-resolved pangenomes to drive marker development, trait discovery, and germplasm characterization
Growing apple-tunities in fruit breeding
Most crop plants have complex genomes characterized by large size, high heterozygosity level and polyploidy.
From the extremely challenging 2.3 Gb maize (85% of which is made up of highly repetitive transposable elements) to the segmental allotetraploid rose and the soybean, which has an astounding 60,000 accessions adapted to different ecoregions, HiFi sequencing has helped scientists not only create more complete blueprints of complex individual lines, but also pangenome collections that capture the genetic diversity of several strains.
We can now add apples to the list.
Recently, Cornell University Orchards in upstate New York provided scientists at the Boyce Thompson Institute and the USDA Agricultural Research Service with source material to build phased, diploid genome assemblies of a selection of domestic and wild apples. Using HiFi reads generated by the Sequel II system, Xuepeng Sun (@XuepengBio), Chen Jiao, Heidi Schwaninger and Zhangjun Fei (@fei_lab), created a pangenome resource that identified genome regions under selection during apple domestication and associated them with important traits such as fruit size, texture and taste.
 And in Nova Scotia, researchers at Dalhousie University are planning to sequence and assemble a haplotype-resolved pangenome from apples grown from one of the most diverse orchards in the world, Canada’s Apple Biodiversity Orchard.
And in Nova Scotia, researchers at Dalhousie University are planning to sequence and assemble a haplotype-resolved pangenome from apples grown from one of the most diverse orchards in the world, Canada’s Apple Biodiversity Orchard.
“We already have a large amount of trait data collected from our orchard of 1,000 apples. Our phenotype data includes traits like firmness, ripening time and sugar content,” said scientist Sophie Watts (@sophia_watts). “The area that is ripe for improvement is our genetic data.”
With resources awarded from the 2021 Plant and Animal SMRT Grant, researchers hope to address several questions with their studies.
Evaluating the genomes of apples from trees that flower and ripen at different times during the growing season could help farmers adjust to climate change, said Zoë Migicovsky (@zoemig).
“As the frequency of unpredictable weather events increases, capturing genetic variation across flowering and ripening time will be an essential starting point for combating the effects of climate change and improving apple resilience,” she added.
HiFi data can help preserve apple biodiversity and better impute complex traits, said Tom Davies.
“High quality reference genomes are like gold for any genetic investigation,” he added. “Full genomes created with HiFi reads will help us to accurately ‘zoom in’ on important areas of the genome that we suspect control important traits, like ripening time and nutritional content.”
Long-read sequencing for spuds
Apples are just one type of produce that can benefit from genomic sequencing. Ever wonder how potatoes grow? As a staple food item for more than 1.3 billion people worldwide, potatoes are a crucial crop throughout the globe. Unfortunately, potatoes are a clonally propagated tetraploid with a large carbon footprint due to pest control, storage, and shipment to farmers. They are also susceptible to many diseases, and historically have not seen genetic gains in breeding.
To address these problems, scientists have invested in efforts to re-invent the potato into an inbred-line-based diploid crop, propagated by seeds.
 Diploid potatoes are possible – in fact, nearly 70% of the natural potato germplasm, including wild species and landraces, are diploid. But scientists trying to create their own diploid hybrids have faced many challenges. Due to deleterious mutations, it has been extremely difficult to develop highly homozygous inbred lines, a prerequisite to breed hybrid potato.
Diploid potatoes are possible – in fact, nearly 70% of the natural potato germplasm, including wild species and landraces, are diploid. But scientists trying to create their own diploid hybrids have faced many challenges. Due to deleterious mutations, it has been extremely difficult to develop highly homozygous inbred lines, a prerequisite to breed hybrid potato.
To overcome these obstacles, an in-depth understanding of potato genomes is required. A team of Chinese scientists led by Sanwen Huang turned to HiFi sequencing to help create a generation of pure and fertile potato lines.
As they detailed in a paper in Cell, they relied on genome analysis for decision making throughout the breeding process, from determining genome homozygosity and the number of deleterious mutations, to haplotype information to infer the linkages between beneficial and deleterious alleles.
The resulting hybrids grew well in both greenhouse and field conditions, with good yield and abundant fruits and tubers rich in carotenoids and dry matter.
The success means that genome-assisted backcrossing, a routine process employed in seed crops, can now be used in potato breeding, with enormous potential to transform potatoes into a more productive crop and a more nutritious food.
“This study transforms potato breeding from a slow, non-accumulative mode into a fast-iterative one, thereby potentiating a broad spectrum of benefits to farmers and consumers,” the authors wrote.
“We even anticipate that hybrid breeding and genome design will transform potato into a non-host to its most devastating disease, the late blight, making it a more environmentally friendly crop.”
Better bovine genomes for breeding
While crops pose a unique set of obstacles, challenges of a different nature emerge when trying to sequence gigabase-sized mammalian genomes in maternal-paternal-offspring trios such as cattle. 
Larger structural variants (SVs) and variations located in repetitive or challenging regions have rarely been studied across Bovinae due to the inherent limitations of short-read sequencing and incomplete reference genomes.
An international team of researchers recently released a pre-print on bioRxiv in which they describe the creation of a cattle pangenome collection.
Researchers examined three bovine trios that reflected diverse breeding strategies (within-breed, inter-subspecies, and inter-species), and created ten haplotype-resolved assemblies that were deemed substantially more contiguous and correct than the current haplotype-merged Bos taurus reference sequence.
The resulting pangenome provided opportunities to study the phylogeny of Bovinae beyond SNPs and indels. The researchers noted that several structural variants were inaccessible from short or noisy long read alignments, such as tandem duplications in QRICH2. Others were challenging to resolve even with good long-read alignments, such as an insertion of HSPA1B.
But, PacBio long-read sequencing methods offered solutions.
“The higher accuracy of HiFi reads was found to be generally more advantageous to quality measures of assembly and increased completeness of centromeric and telomeric regions
compared to the longer but higher-error ONT reads,” they wrote.
In this case, the accuracy difference was clear.
“Shasta assemblies averaged QV 41.5 after one round each of polishing with ONT reads and short reads, while hifiasm assemblies reach QV 47.6 without any polishing” the authors observed.
They went on to say “…hifiasm assemblies had a 4-fold reduction in base errors compared to the Shasta assemblies, indicating the ability of HiFi data to achieve higher quality in fewer steps.”
HiFi-based assemblers required less compute and storage resources, they noted. Producing haplotype-resolved HiFi assemblies required approximately 600 CPU hours and 200 GB of peak memory usage, while the equivalent ONT-based Shasta (plus polishing) assemblies took 2200 CPU hours and 750 GB of peak memory usage.
To learn more about the bovine genome assembly project, check out this video presentation by Tim Smith of the USDA’s Agricultural Research Service, or this SMRT Leiden talk by Hubert Pausch of ETH Zurich.
Check out our new brochure, or see additional examples of the use of HiFi Sequencing for the generation of plant and animal pangenomes:
Webinar Summary: Crops and Corvids get the Pangenome Treatment with HiFi Sequencing
Pangenome of Soybean Generated to Capture Genomic Diversity
Project to Rapidly Sequence Maize Pangenome Delivers Publicly Available Resource
Sequencing 101: Looking Beyond the Single Reference Genome to a Pangenome for Every Species
Case Study: Pioneering a Pan-Genome Reference Collection