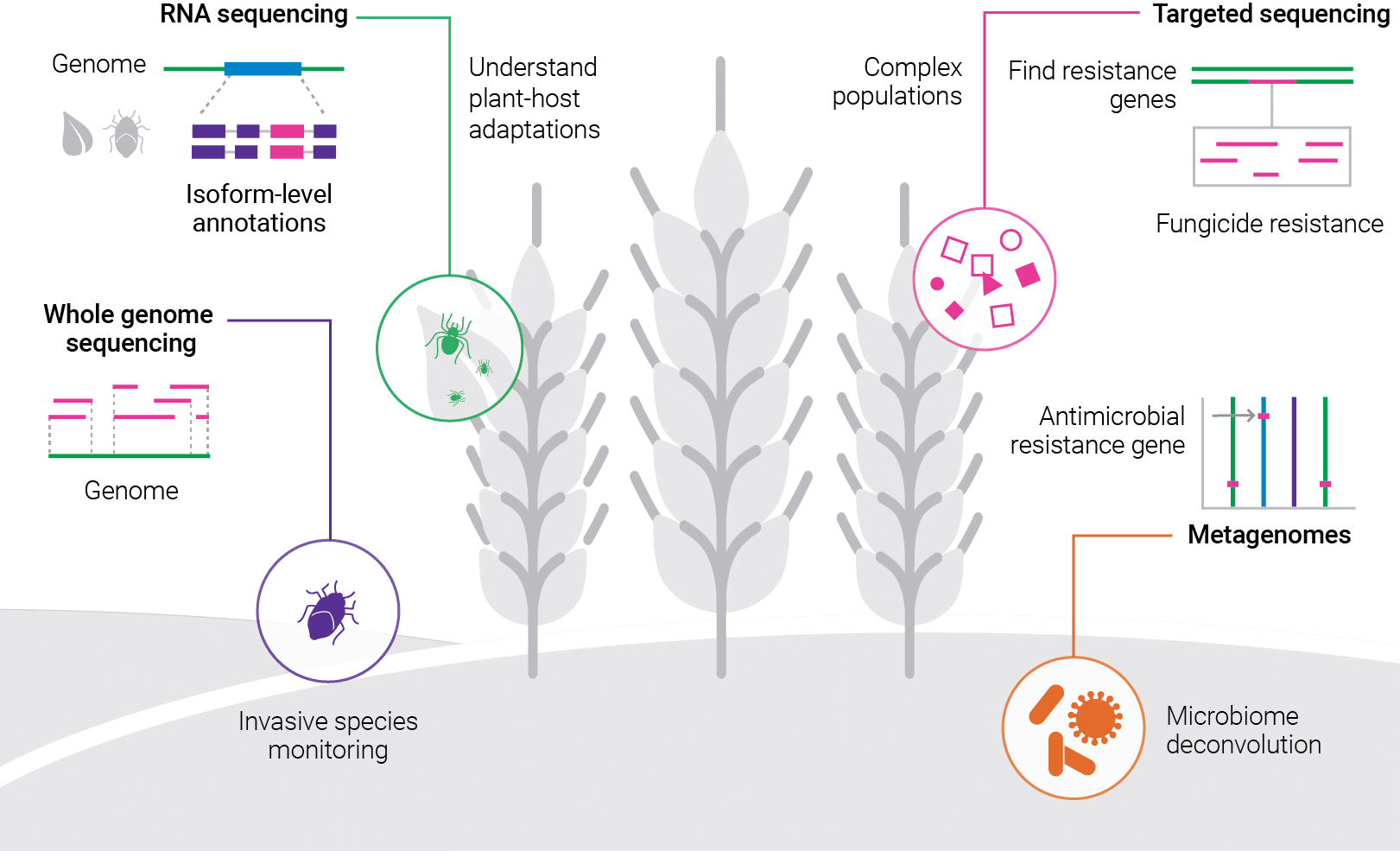
 When you’re dealing with insects, fungi and microbes with short life cycles and rapid reproduction, you often encounter highly variable genetics within single populations. The complexities of genetics within these populations are one of many challenges faced by researchers and field scientists studying invasive species and agricultural pests.
When you’re dealing with insects, fungi and microbes with short life cycles and rapid reproduction, you often encounter highly variable genetics within single populations. The complexities of genetics within these populations are one of many challenges faced by researchers and field scientists studying invasive species and agricultural pests.
Luckily, HiFi sequencing has emerged as a handy tool to help researchers find novel genes that are responsible for immunity, metabolic detoxification, and resistance to pesticides in insects, fungi, and microbes.
Using HiFi sequencing to assemble reference genomes of pesky pests
The USDA’s Agricultural Research Service utilizes HiFi sequencing to develop low-cost, high-quality reference genomes from single insect specimens of all sizes, and the development of these reference genomes has evolved over time.
When the Ag100Pest Initiative was initiated in 2018, only 6 of 366 (1.6%) arthropod genomes available through NCBI met high standards of contiguity. As the name implies, the consortium originally intended to generate reference genome assemblies of 100 pests or potential pests, but the project grew over time and now a total of 158 species are currently in the pipeline. This includes 18 families and 121 species that lack a publicly available assembly of any quality.
Insects are not a new threat to agriculture, but their impact on production have been greatly affected by pesticide use, climate change, and the introduction of non-native insects into new habitats through global trade. Widespread insecticide resistance has emerged, expanded seasonal activity of native pests have increased damage to geographic landscapes, and the migration of non-native pests between habitats has challenged ecosystems.
“Our ability to control arthropod pests must undoubtedly also evolve and adapt to mitigate these threats, and genomics, in particular, holds promise to facilitate the development of innovative and resilient control technologies,” Ag100Pest leaders wrote in a 2021 paper.
It is clear that high-quality genome assemblies are an integral part of their work.
“Recent estimates suggest that nearly 90% of economically or ecologically important traits in organisms may be determined by variation in non-coding regions of the genome indicating the need for high-quality reference genome assemblies to study traits relevant to pest management,” they wrote.
The authors specifically called out PacBio’s standard input protocols and the accuracy of HiFi reads as “key to the success of the Initiative.” By lowering DNA input requirements, PacBio’s new workflows are more accessible than ever for obtaining DNA for even the smallest of organisms.
The benefits could be vast, including supporting the bioeconomy through technological advances in the efficacy and durability of environmentally sustainable pest management practices, such as novel molecular-based management tools that target pests while sparing environmental damage, particularly damage to beneficial arthropod populations.
“Increasing profitability while reducing any negative environmental impacts of agricultural production directly benefits rural economies, societal well-being, and overall human health,” they conclude.
PacBio assays in action around the world
With increasing restrictions surrounding the registration and usage of pesticides in the European Union, it is essential to ensure existing fungicides remain effective. Understanding the dynamics of fungicide resistance in target pathogen populations is therefore a key pillar of integrated pest management programs around the world.
In some cases, pesticide and fungicide resistance is made possible through target-site mutations, with pathogens adapting by accumulating multiple target-site alterations. Monitoring resistance through the molecular detection of alterations in the target site genes can be a useful tool.
A team of researchers in Ireland, Sweden, Denmark, and Belgium has taken this approach in their study of Zymoseptoria tritici, which causes Septoria tritici blotch (STB), the most destructive disease of winter wheat throughout western and northern Europe.
As they describe in this Frontiers in Microbiology article, the researchers created a PacBio long-read sequencing assay built on multiplex amplification of target genes to enable the simultaneous detection of alterations across several areas which are suspected of playing a role in enabling resistance in Z. tritici to three types of fungicides. The pooled PCR products from two multiplex PCRs (containing 14 different amplicons from 96 isolates) were sequenced in one SMRT Cell.
The researchers also included multiplex amplification and sequencing of nine housekeeping genes alongside those associated with fungicide resistance to provide comprehensive Z. tritici strain genotyping. The assay enabled them to advance their understanding of the pathogen’s biology, and its potential utility for future applications.
“When utilized to its full capacity to screen multiple targets in large collections of isolates, costs per strain can be expected to be considerably lower compared to traditional Sanger sequencing,” they concluded.
Insights from Iso-Seq analysis
Still another way creatures can gain resistance is via alternative splicing, a common feature in eukaryotes that increases the transcript diversity with functional consequences.
In insects, alternative splicing has been found associated with resistance to pesticides and Bt toxins, and researchers in Missouri were curious to see whether it played a role in Bt resistance in the western corn rootworm (Diabrotica virgifera virgifera LeConte), which causes an estimated $1.4 billion in damage to US corn production each year.
As reported in the Royal Entomological Society’s journal Insect Molecular Biology, the team carried out Single Molecule Real-Time (SMRT) transcript sequencing and Iso-Seq analysis on western corn rootworm neonate midguts which fed on seedling maize with and without the chimeric protein present in transgenic maize (Cry3.1Ab).
Using a combination of RNA-seq and Iso-Seq transcript sequencing techniques, they analyzed transcriptome-wide alternative splicing patterns of the rootworm midgut in response to feeding on eCry3.1Ab-expressing corn.
Our planet is an intertwined series of ecosystems that house microbes, plants, and animals. Each part of these ecosystems play their part in supporting the planet and the lives and wellbeing of those who inhabit it. To better understand invasive species and protect our crops and livestock from pests and disease, researchers need reference genomes that they can count on — and Hifi sequencing can help.
Interested in learning more about using HiFi sequencing in insect and pest research? Check out our Plant and Animal page.
Or, read the other posts in this series:
Agrigenomics for crop and livestock breeding
Agrigenomics for genome engineering