We’re thrilled to have recently launched our summer promotions for Onso sequencing. These offers open the door for researchers to experience the incredible accuracy of Q40+ accuracy, at a cost that’s lower than lesser performing systems.
Why is this so exciting? Achieving Q40+ accuracy in your sequencing means unlocking new levels of precision and certainty in your research. For over a decade, the Q30 standard of next-generation sequencing accuracy has been considered “good enough,” despite its inherent limitations. The introduction of Onso short-read sequencing redefines this standard, offering extraordinarily accurate reads with an error rate of only 1 in 10,000 bases or less (a quality score of Q40+).
This extraordinary accuracy is achieved through innovative sequencing by binding (SBB) chemistry, which includes several pivotal enhancements over traditional sequencing by synthesis (SBS) chemistry. High SBB accuracy provides optimized sequencing chemistries, lack of molecular scarring, and delivers extremely low false positives leading to greater sensitivity for rare variant detection.
We’ve already shown you how the Onso system excels at needle-in-a-haystack applications, like liquid biopsy research. Now, explore how the exceptional accuracy of the Onso system extends beyond the proverbial haystack across diverse fields such as oncology, public health, and human genetics research, illustrated by three insightful case studies.
Keeping closer tabs on more bacterial species in public health monitoring
While it first made headlines during the Covid-19 pandemic, wastewater surveillance has become a routine and indispensable tool for epidemiology and public health. Some of the credit for this evolution can be attributed to modern culture-independent sequencing.
PacBio and Zymo Research recently collaborated on a wastewater surveillance study, presented at this year’s ASM meeting. This study used Zymo’s Quick-DNA/RNA Water Kit to process real-world wastewater samples. They were then sequenced on the PacBio Onso system and the Illumina NextSeq 2000 System in a head-to-head comparison of performance for pathogen detection, antimicrobial resistance (AMR) monitoring, and functional analysis.
Across all samples, sequencing on the Onso system with the Zymo analysis workflow generated more taxa for each taxonomic level compared to the NextSeq 2000 System. That means that more microbial diversity was detected with PacBio SBB than SBS (Figure 1).
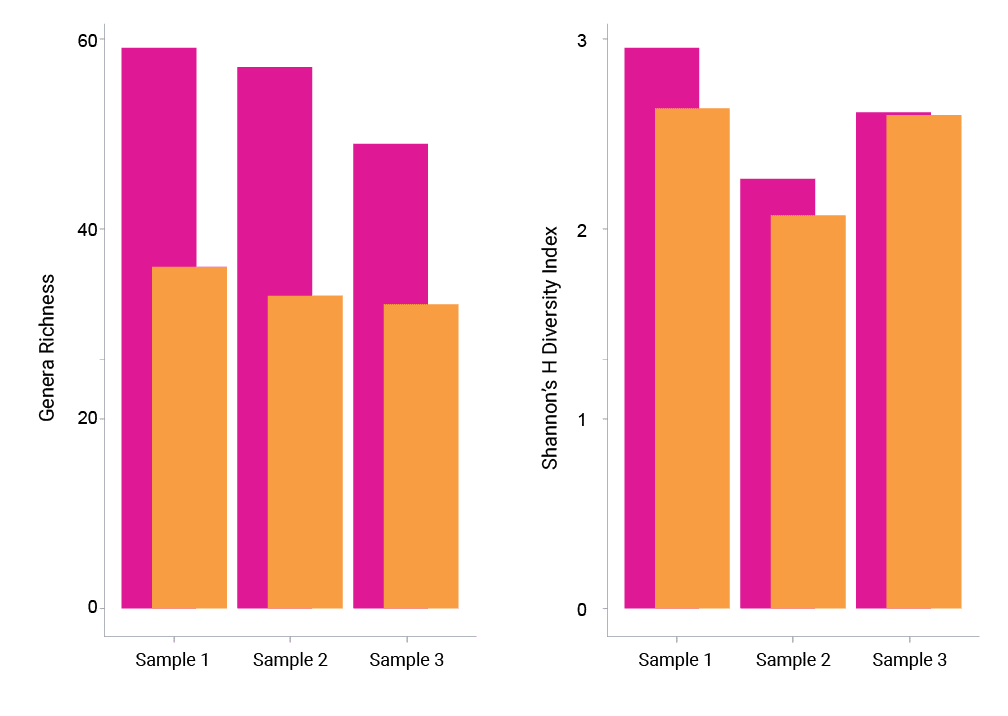
SBB on the Onso system also detected more antimicrobial resistance genes than the NextSeq 2000 System (Figure 2). This outperformance is largely due to the fact that the Onso system is capable of higher sensitivity of detection than the competition.
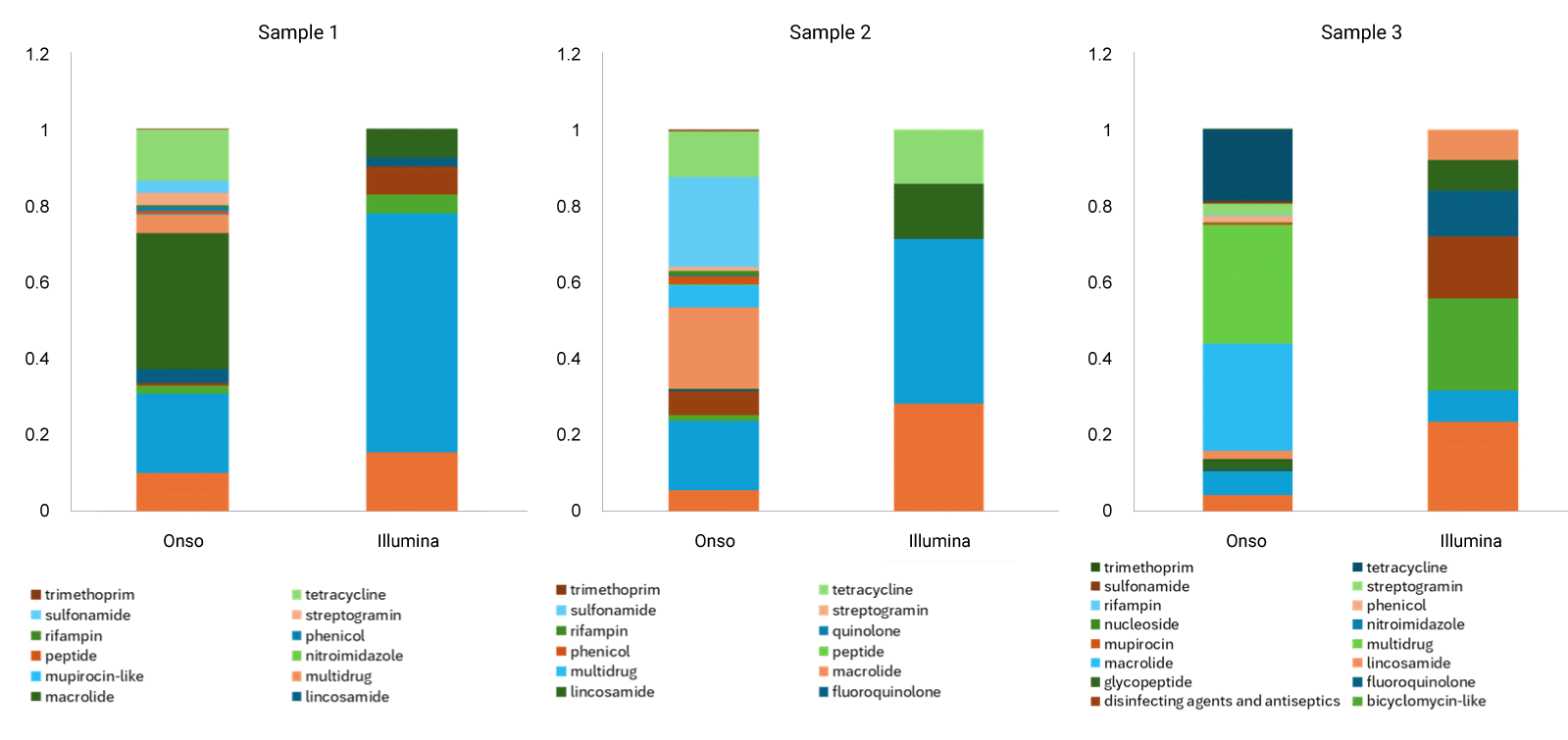
Although epidemiology is quite different from oncology research, both fields share notoriously challenging sample types and a need for high sensitivity and specificity, while human health is at stake. The Onso system demonstrates that better detection means better results.
The high accuracy of SBB equips researchers with the tools necessary for monitoring and predicting outbreaks, optimizing public health management and water treatment strategies, and making recommendations for better public health policies.
Lower duplication rates mean fewer wasted reads for germline variant detection
Resolving clinically actionable information demands high accuracy. Furthermore, as sequencing capabilities have grown, clinical sequencing has shifted from more targeted regions to whole exomes and whole genomes. This shift means that the highest accuracy sequencing is not only desirable but an absolute necessity to avoid errors in clinical information that can impact therapy decisions and deeper understanding of diseases.1
Another collaboration, this time between PacBio and Twist Bioscience, demonstrates how SBB on the Onso system delivers yet again when accuracy matters. In this study, 8 replicates of NA12878 gDNA were fragmented using Twist’s Enzymatic Fragmentation 2.0 kit. Regions of interest were captured with the Twist Exome 2.0 panel and existing P5/P7 libraries were converted using the Onso library conversion kit. The researchers then performed whole-exome sequencing on the Onso system using 2 x 100 bp reads and compared it to SBS.
Their findings show that whole-exome sequencing on the Onso system exceeded system specifications for reads, output, and accuracy (Table 1).
Table 1
| Metric | Spec | Run 1 | Run 2 |
|---|---|---|---|
| Total reads | 800 M | 1,062 M | 1,107 M |
| Read length | 2 x 100 | 2 x 100 | 2 x 100 |
| Output | 80 Gb | 116 Gb | 119 Gb |
| Accuracy (90% Q score) | 40 | 49 | 44 |
They also found that SBB on the Onso system performed comparably to or better than SBS when measuring the percentage of bases over 30x coverage, uniformity of coverage, and duplication rate (Figure 3). This effect was even more pronounced in medically relevant regions, represented by ACMG 81 (a list of 81 genes containing pathogenic or likely pathogenic variants). The lower duplication rate with SBB translates to fewer wasted reads overall vs SBS.
Overall, these results highlight the value SBB on the Onso system brings to variant detection in clinical exome sequencing and disease research. For more details, you can explore the public dataset.
High sensitivity and specificity for variant detection in cancer research
As shown, liquid biopsy applications are a promising noninvasive method for both basic research and potential diagnostic research, but NGS-based analyses of these samples are challenging due to the low concentration of cell-free DNA (cfDNA). Traditional short-read sequencing technology has struggled to capture these rare pieces of DNA and sequence them with the sensitivity and specificity needed for reliable detection.
A collaboration between PacBio and IDT demonstrates how the Onso short-read sequencer successfully detected low-frequency variants in acute myeloid leukemia (AML) samples. This study used IDT xGen high-conversion library prep technology to prepare libraries from AML cfDNA and fragmented gDNA reference samples.
The collaborators tested this workflow with DNA inputs ranging from 1 ng to 50 ng cfDNA to mimic the small amounts of DNA found in real patient samples. All libraries, even the low input of 1 ng, had >99.9% of reads aligned to the reference and ≥75% of bases on target, which is particularly impressive given the small panel size (~9 kb).
When sequenced on the Onso system, the converted xGen libraries yielded extraordinary accuracy: 96% of bases at Q40+ and 83% of bases at Q50+. This level of sequencing accuracy is unheard of on other short-read sequencers, like SBS systems. Even for inputs as low as 1 ng, the Onso system delivered >99.5% sensitivity and ≥99.7% specificity at 250K reads/sample for the gDNA reference samples (Figure 4), and performance was maintained even at very low subsampling depths (<24K reads/sample). Performance for the cfDNA reference samples was similarly impressive, achieving 100% sensitivity and ≥99.7% specificity even for variant allele frequencies (VAFs) as low as 0.5%.
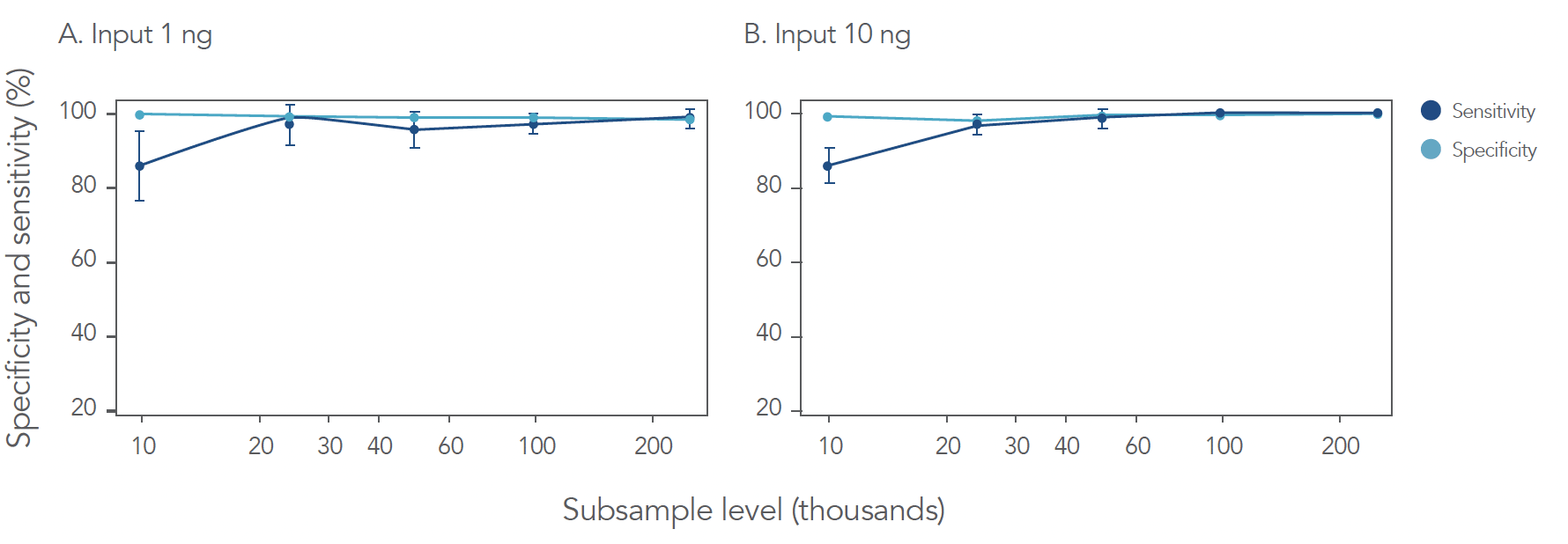
SBB accuracy and low noise enables superior sensitivity and specificity across a 10-fold range of read depth. These results demonstrate the possibility of adjusting sequencing depth to achieve experimental goals – a refreshing change from existing approaches where deeper sequencing depths are the norm for somatic variant detection.
To see the full details of this study, read the technical note or explore the dataset.
Boldly going beyond the haystack
These studies are only a few examples of the potential applications of the Onso system, where accuracy continues to matter beyond the “needle in the haystack.” This next level of accuracy can empower public health, disease research, and more. There’s no limit to what you can achieve in this era of accuracy.
Ready to make the switch to Onso?
Trade up your tired old Q30s for the possibilities of Q40, Q50, and beyond. Get the Onso system for just $99k with the trade-in of any non-PacBio NGS system.
Reference:
- Poplin R, Zook JM, DePristo M. Challenges of Accuracy in Germline Clinical Sequencing Data. 2021;326(3):268–269. doi:10.1001/jama.2021.0407