
Highly-accurate HiFi reads produced by PacBio long-read sequencers –such as the Sequel IIe and Revio systems– enable researchers to access entirely new levels of contiguity, completeness, and accuracy when assembling genomes.
The sequencing capabilities of these long-read platforms are particularly beneficial for microbial genome assembly. The superiority of PacBio assemblies enables microbiologists working on the front lines of discovery to investigate animal and plant health, foodborne illnesses, and antimicrobial resistance more comprehensively and with greater accuracy and detail than ever before. Even so, methods upstream of sequencing –such as DNA extraction, shearing, and library preparation– have proven challenging to users looking to scale their microbial long-read sample pipelines. Workflow speed and cost-effectiveness has limited the use of HiFi microbial whole genome sequencing (WGS) data in some public health and infectious disease settings –until now.
A high-throughput solution for laboratories great and small
In collaboration with a customer that specializes in generating high-impact genomic data at scale, PacBio has introduced a revolutionary set of materials and protocols that can accelerate and economize the sample-to-data process for nearly any PacBio long-read user, achieving impressive throughput enhancements of 4 to 12-fold. Key benefits at the shearing step of the workflow include:
- 3-minute processing time
- Up to 96 samples per plate
- <$1.00/sample
These new protocol options enable researchers to attain a more streamlined extraction, shearing, and library preparation experience for PacBio long-read sequencing applications –regardless of project size. The workflows also include a newly implemented assembly algorithm for HiFi reads that works directly within SMRT Link, the PacBio software that is used to set up and run sequencing experiments. Combined, these new workflows and software enhancements give microbiologists access to a single high-throughput, end-to-end solution for microbial genome assembly.
Let’s take a quick look at how this new protocol is set up.
How the rapid HiFi microbial WGS workflow works
The new end-to-end workflows include a few streamlined steps to complete. This approach can be closely reproduced with a variety of extraction kits –such as the Nanobind HT CBB kit– using instruments from Thermo Fisher and Hamilton and can serve as a starting point to suit individual needs.
Here is a brief overview of these high-throughput approaches, with valuable recommendations and key figures highlighting performance:
Workflow outline
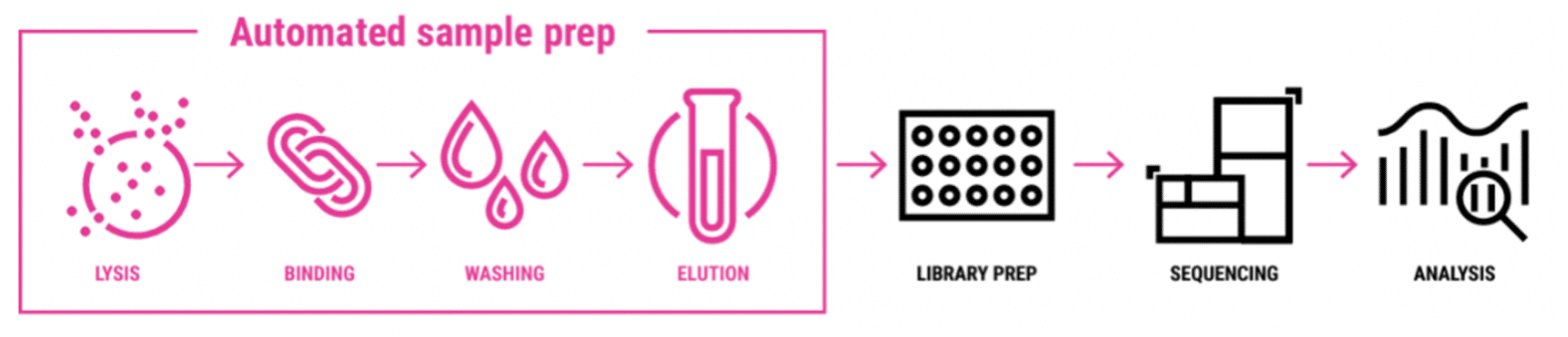
- Extract DNA using the high-throughput (HT) protocol.
- Start with the recommended input of 300 ng high-quality DNA.
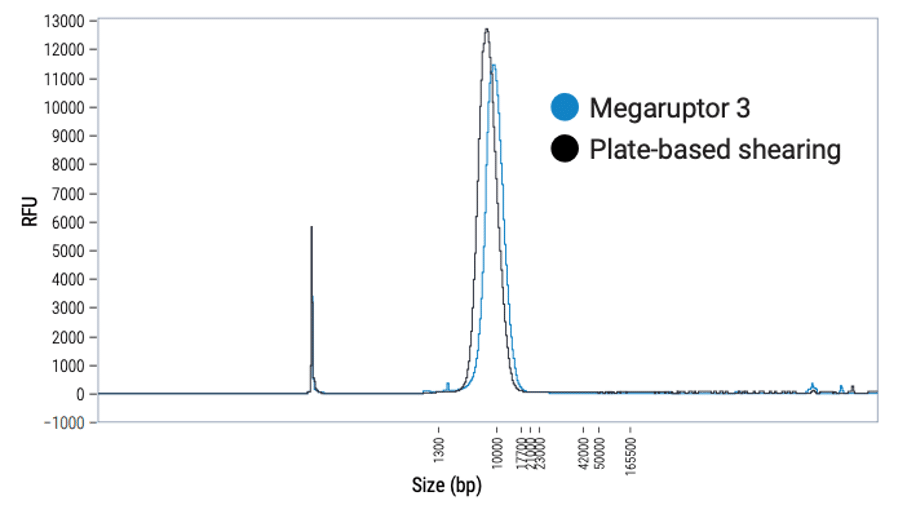
- Shear DNA to an average size of 7–10 kb using the plate-based high-throughput method.
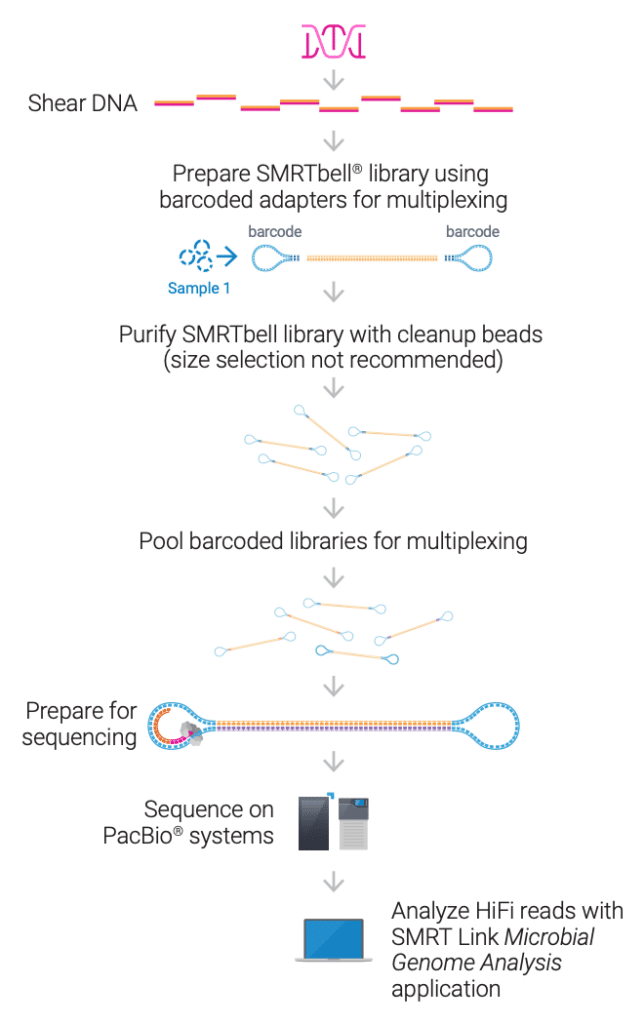
- Prepare libraries using the SMRTbell prep kit 3.0 (PN 102-182-700) with the SMRTbell barcoded adapter plate 3.0 (PN 102-009-200) which enables multiplexing up to 96 samples. Automated protocol available from Hamilton.
- Load and run samples on PacBio long-read sequencing system.
- Use SMRT® Link for fully automated de-multiplexing, assembly, circularization, and polishing of both chromosomes and plasmids to produce reference-grade assemblies of outstanding quality and completeness.
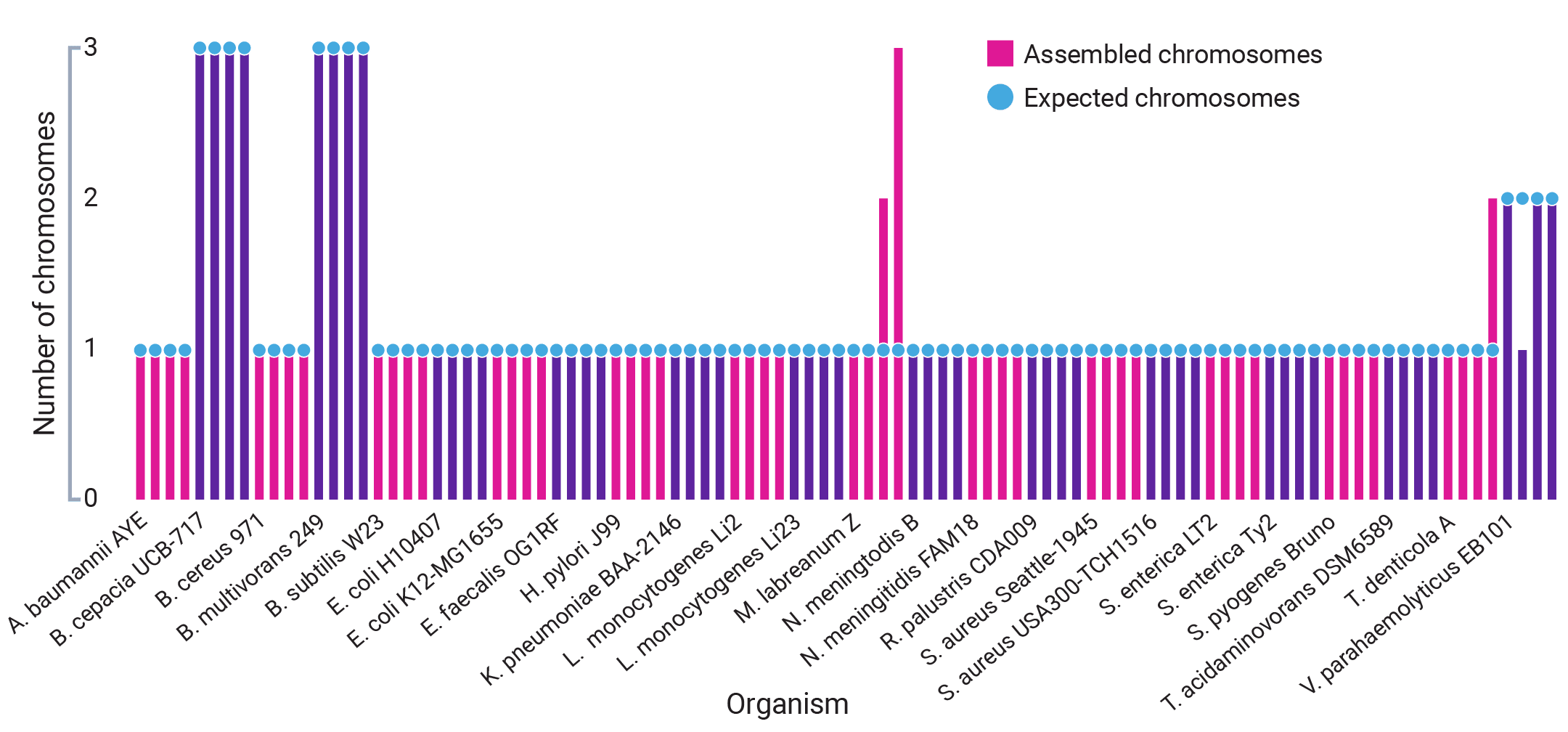
- Achieve high-quality consensus accuracies >99.99%
- Output data in standard file formats (BAM and FASTA/Q) for seamless integration with your preferred downstream analysis tools.
Changing the game in microbial genomics and beyond
The ongoing collaborative development of high-throughput workflows like this make HiFi microbial WGS more cost and time efficient than ever before.
While these protocols open many new doors for microbial WGS, the core principles of what we’ve optimized –such as plate-based shearing– can be easily adapted to a host of other sequencing applications to dramatically increase efficiency. These include HiFi sequencing across diverse domains—plants, animals, humans, metagenomics, and more. In today’s genomics landscape, it’s imperative to empower researchers to populate reference databases with top-tier genomes. This access accelerates taxonomic and functional classification, and even bolsters objectives like pandemic preparedness. HiFi sequencing sets the stage for this to become the norm, enabling you to shape a new standard of excellence in microbial genomics.
Want to learn more about PacBio sequencing?
Try out PacBio data
We encourage you to download some datasets and experience the impressive quality of HiFi data for yourself.
Explore these resources to help initiate your project
We have tons of information on how to get started with PacBio sequencing. Check out these resources to see how:
Microbial WGS best practices guide
Automating library prep using Hamilton instruments
HT plant + microbial sequencing workflows
HT shearing technical note
Public health brochure
PAG 2023 poster
APHL/ASM Microbe 2022 poster