In this article, we discuss PacBio RNA sequencing through the lens of our most recent Iso-Seq Social club event. Read about the exciting scientific wins that were shared, and see how the all-new MAS-Seq protocol is being applied.
Background on the Iso-Seq method
With long read lengths and immaculate detail, HiFi sequencing is the gold-standard for creating reference-grade assembles of even the most challenging genomes. The PacBio Iso-Seq method brings the explanatory power of HiFi sequencing to the world of transcriptomics, enabling impactful and groundbreaking research based on a complete view of isoform diversity.
When applied to single-cell RNA studies, HiFi sequencing combined with the all-new 10x compatible MAS-Seq method greatly increases throughput to deliver insights that are out-of-reach with conventional short-read approaches.
Advantages of the Iso-Seq method include:
- Full-length reads that capture the entire transcript, including untranslated regions
- Alternative start- and end-site detection
- Characterization of splicing events
- Allele-specific isoform identification
- Expression profiling at isoform resolution
- Fusion gene detection
- And so much more!
Learn more about the Iso-Seq method
An event for accelerating discovery through the exchange of ideas
Whether you are new to the Iso-Seq method, or are a seasoned veteran of the technology, Iso-Seq Social Club events are the place to tap into the creativity of the user community and invigorate your own research.
The Iso-Seq Social Club was originally devised to keep the energy of the die-hard Iso-Seq community alive during the height of the global COVID-19 pandemic. Today it has grown well beyond its humble roots into a dynamic digital event with a multitude of talks from passionate scientific professionals across academia and industry.
The Iso-Seq Social Club provides attendees a rare opportunity to engage with topics from a range of life science fields unified by a common research methodology –an ideal environment for spontaneous insights and cross-disciplinary methodological innovation.
Topics covered in the most recent Iso-Seq Social Club event (Vol.3) include:
- An introduction to the Iso-Seq method
- SQANTI3 for isoform classification and annotation
- TappAS for isoform differential expression analysis
- Bulk Iso-Seq applications: from disease to function
- Bioinformatics tools for Iso-Seq and single-cell Iso-Seq analysis
- Single-cell Iso-Seq applications in cancer and neurological disorders
Watch the talks from Iso-Seq Social Club Vol. 3 on-demand
Big moments from Iso-Seq Social Club Vol. 3
Swaths of novel isoforms discovered in preimplantation embryos
Denis Torre, a PhD candidate in the Sebra and Guccione labs at the Icahn School of Medicine at Mount Sinai, presented his work on gene expression during the complex process of human embryonic development in the preimplantation phase. Using 73 embryos between the 1C and blastocyst stages, Mr. Torre and collaborators used the Iso-Seq method to uncover more than 100,000 novel mRNA isoforms including numerous novel-not-in-catalog, novel-in-catalog, antisense, and intergenic sequences.
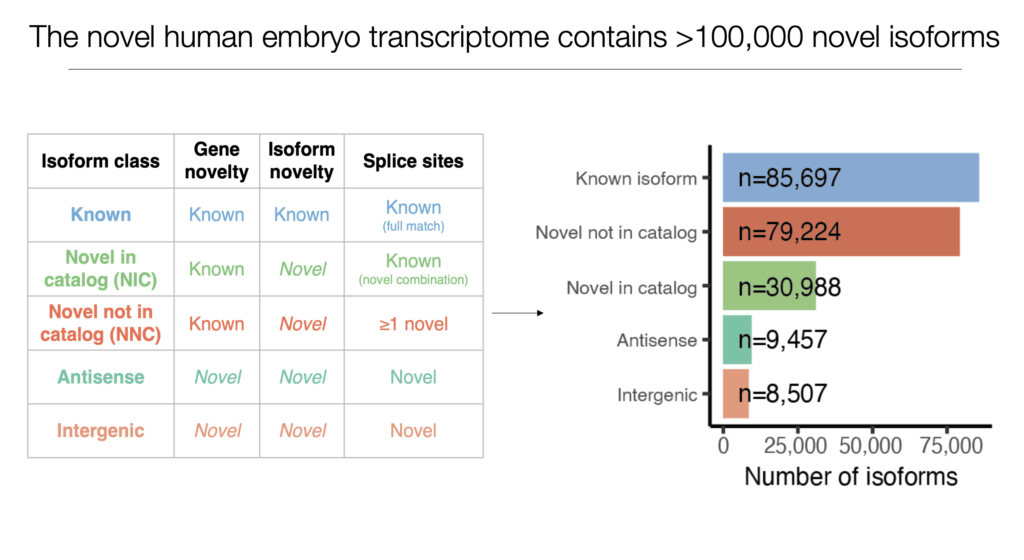
Using a combination of long and short-read RNA-seq data Torre and collaborators then moved to classify these novel isoforms and tease apart some of their fundamental characteristics. What they found was that the various isoform classes displayed differences in protein-coding potential, inclusion of repetitive elements, and evolutionary conservation. They also observed that most novel gene and isoform expression takes place in the early stages of preimplantation, after which they are downregulated.
Unique splicing patterns and new protein isoforms revealed with long-read proteogenomics using Iso-Seq data
Madison Mehlferber, a PhD candidate in the Sheynkman Lab at the University of Virginia presented her work on the development of a systematic method to enhance the characterization of protein isoforms with long-read proteogenomics. Using human umbilical vein endothelial cells (HUVECs) as a model system, Mehlferber was able to overlay peptide sequencing information with Iso-Seq data to infer the presence of novel protein isoforms in cases where sequence database information was limiting. This approach was further used to estimate relative protein isoform abundances with promising results. Future research directions for Mehlferber and the Sheynkman Lab include applying this approach to the study of developmental changes and disease models.
Exciting developments in single-cell transcriptomics
This year’s event included talks on methodological innovations that massively increase throughput and data quality for single-cell analyses as well as promising applications in translational research. Speaking from the analysis side, Roger Volden, PhD, a bioinformaticist at PacBio opened the informatics session with a walk-through of PacBio SMRT Link v11.1. This software release offers PacBio users a push-button workflow for processing data from MAS-Seq, a new kitted solution for single-cell isoform analysis that leverages molecular concatenation to turbocharge transcript capture by 16x. In a similarly themed talk, Jon Armstrong of Jumpcode Genomics presented a method developed by the company that boosts isoform recovery even further by using CRISPR-Cas9 to deplete cDNA libraries of rRNA and other unwanted molecules that contaminate sequencing data.
On the translational side, Katie Miller, PhD, a researcher at Nationwide Children’s Hospital presented her group’s work on somatic mosaicisms and somatic mutations associated with pediatric epilepsy. Through earlier studies, the team was able to identify two pathogenic somatic variants observed in PTEN. However, due to the limitations of short-read sequencing they were unable to properly phase and discern whether the mutations occurred on the same allele. Using a targeted bulk Iso-Seq approach, Miller’s team was able to leverage the length and accuracy of HiFi sequencing to determine that the variants did not occur on the same allele, a critical insight for guiding further analysis. Taking the granularity of the study one step further, single-cell Iso-Seq was used to overlay gene expression onto genotype to reveal the cell types carrying these mutations, which were revealed to be oligodendrocytes and astrocytes. Dr. Miller also shared another study where RHEB was genotyped using the new MAS-Seq approach.
Watch to these talks on-demand to find out more
Check out additional Iso-Seq resources and upcoming events
At PacBio, nothing matters more than being a reliable partner who supports your scientific success. Check out these additional resources and opportunities to discuss how PacBio can help turn your research vision into an actionable and impactful reality.
- Attend the upcoming MAS-Seq webinar On November 16, 2022, at 1:00PM EST PacBio scientists will present the MAS-Seq method, an all-new kitted single-cell transcriptomics solution that delivers unmatched data quality and game-changing explanatory power for single-cell research.
- Watch Iso-Seq Social Club Vol. 3 talks, on-demand
- Learn more about the Iso-Seq method
- Connect with a PacBio scientist to discuss your RNA experiment