
A benchmarking of long-read RNA sequencing methods and analysis tools
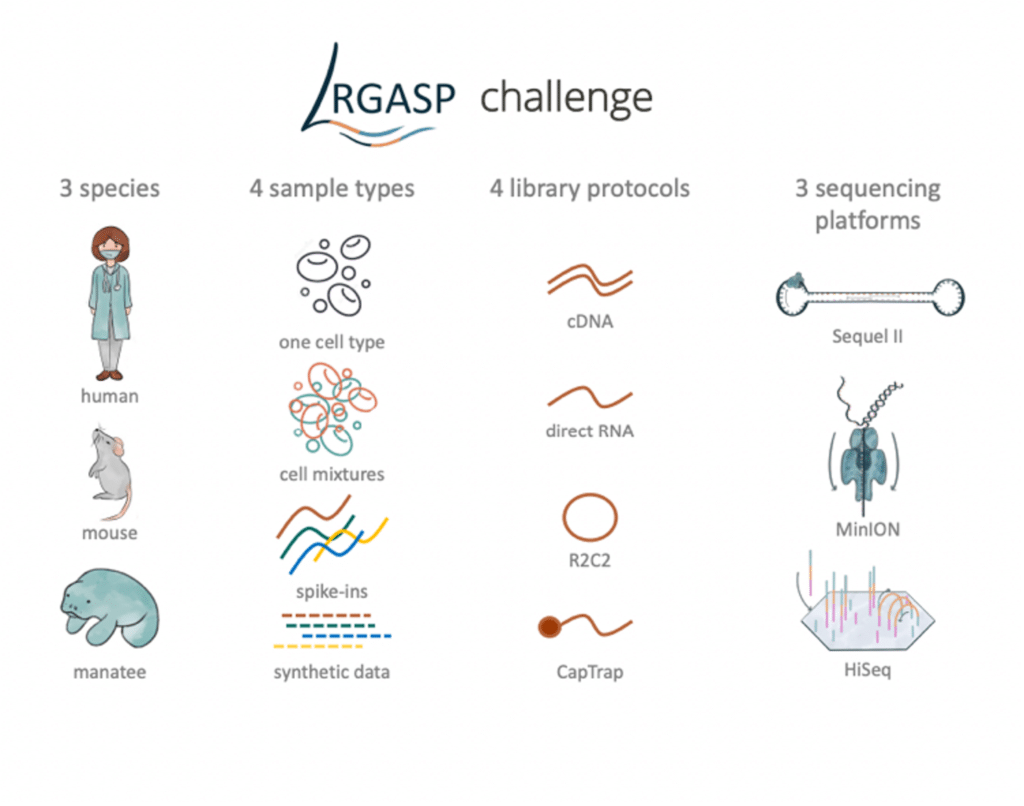
The Long-read RNA-Seq Genome Annotation Assessment Project (LRGASP) consortium, an initiative to systematically evaluate methods for transcript identification and quantification, recently released their final assessment of long-read sequencing technologies and tools in the preprint “Systematic assessment of long-read RNA-seq methods for transcript identification and quantification”. The consortium collected sequencing data on three different species, cell types and mixtures including synthetic spike-in RNA controls, different library preparations, and sequencing platforms (Figure 1). Across all platforms and samples, the consortium generated a total of >400 million reads.
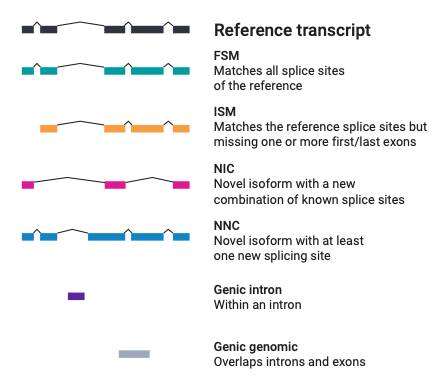
To assess the quality of the RNA sequencing data generated by different library preparation, sequencing methods, and software tools, the consortium used SQANTI3 – a long-read isoform classification and QC software tool. SQANTI3 compares long-read isoforms against existing annotations (e.g. GENCODE) to characterize them as known or novel genes or isoforms, as well as using orthogonal information to evaluate the 5’ and 3’ completeness of the transcripts.
Iso-Seq method detects more long and rare isoforms accurately
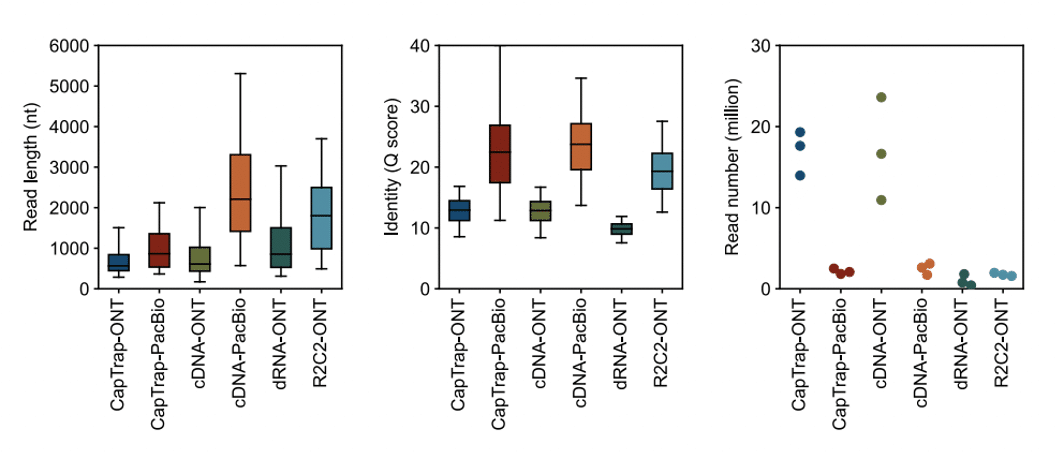
The consortium found that PacBio sequencing detected the greatest number of genes. While different long-read RNA-Seq tools varied greatly in the number of isoforms detected, “cDNA library preps, especially in combination with PacBio, more frequently had the highest number of FSM, NIC, and NNC isoforms”. Meanwhile, the consortium found that Oxford Nanopore (ONT) data more frequently included anti-sense and genic genomic transcripts, which are likely to be library artifacts.
In addition, using PacBio with the standard Iso-Seq library preparation “was the experimental procedure where their exclusively detected transcripts were the longest and had significantly lower expression”. This is despite having collected more ONT data for the LRGASP study compared to other technologies, where the authors observed “more reads did not consistently lead to more transcripts, indicating that read quality and length are important factors for transcript identification.” The ability to capture long and real transcripts was further confirmed with the SIRV (RNA spike-in) controls, where the PacBio Iso-Seq method was the only method that recovered all SIRV transcripts. In contrast, the CapTrap method (a cDNA library preparation method that combines Cap-trapping with oligo-dT priming to capture 5’ capped transcripts) using PacBio sequencing showed limitations in capturing long molecules.
Iso-Seq method is more accurate for transcript quantification
While PacBio and ONT cDNA libraries both had good reproducibility and consistency across replicates, the consortium found PacBio Iso-Seq method to have 2-fold higher abundance resolution (ability to quantify isoforms) compared to ONT cDNA data. This is further supported by PacBio Iso-Seq method’s outperforming other methods in the SIRV synthetic spike-in data for isoform-level quantification. Overall, the consortium found RSEM to be the most consistent software tool for quantifying long-read RNA-Seq data across a variety of platforms and conditions, while IsoQuant, IsoTools and FLAIR were also well-performing.
The consortium pointed out that with the increase in throughput for long-read RNA-Seq methods, “the quantification accuracy of long-read-based tools is likely to be further improved.”
Experimental validation of novel isoforms shows the power of long-read RNA-Seq
The consortium validated novel isoforms discovered using PacBio and ONT data with targeted PCR and obtained a 100% validation rate for novel isoforms that were consistently detected across software pipelines. More surprising was the high validation rate of isoforms with exceedingly low reproducibility across software pipelines. These validation experiments underscore that novel isoforms found by PacBio and ONT likely capture biologically real isoforms. “Novel isoform predictions generally have high accuracy, even if such isoforms are not consistently predicted across pipelines and platforms,” the consortium writes, while isoform validation success is most related to how often the isoform is detected or how abundant it is.
Looking to the future: scaling up long-read sequencing with MAS-Seq for bulk Iso-Seq method
The LRGASP consortium study highlights the value of PacBio Iso-Seq data for accurately detecting long, novel and rare transcript isoforms and quantifying them across different samples. A noted limitation was the lower throughput of Iso-Seq data using the Sequel II system. With the throughput increase of the MAS-Seq concatenation method along with the higher throughput Revio system, MAS-Seq for bulk Iso-Seq will greatly reduce the cost of high-quality full-length isoform sequencing.
Learn more about how researchers are using the PacBio Iso-seq method and MAS-seq by registering for the upcoming Long-read RNA Neurology Symposium, or visiting us at ASHG 2023.