RNA sequencing (RNA-seq) has become an indispensable tool for analyzing transcriptomes across all domains of life that can reveal insights about biology and disease. While the genome remains relatively constant for most species over brief time scales, the transcriptome — the sum total of expressed messenger RNA (mRNA) transcripts — varies by many factors, including developmental stage, ecological conditions, and disease. However, not all sequencing technologies are equal when it comes to RNA sequencing.
In this blogpost, we will walk through a brief history of full-length RNA-seq using PacBio HiFi sequencing, review the previous limitations on throughput, and the groundbreaking promise that the recently launched Kinnex full-length RNA kits brings.
Isoforms, not genes, are the drivers of biology and disease
Sometimes scientific challenges require innovative technologies to bring about new discoveries. Long-read RNA sequencing using PacBio HiFi (referred to as the Iso-Seq method) is dramatically different from traditional short-read RNA-seq. Instead of fragmenting full-length cDNA (which can range up to tens of kilobases) into short 100–200 bp fragments that are then computationally assembled, the PacBio Iso-Seq method sequences the entire full-length molecule; no assembly required.
The Iso-Seq method is advantageous because alternative splicing in eukaryotic species can produce multiple complex messenger RNAs (a.k.a. isoforms) from the same gene. Different isoforms can then be translated into different proteins that have distinct, and sometimes opposing, functions. The study of biology and disease through the lens of RNA sequencing therefore must have a precise and exacting view of the isoform landscape. Short-read RNA-seq often cannot unambiguously resolve complex alternative splicing events, whereas the Iso-Seq method is able to reveal them clearly without any of the same computational challenges.
The Iso-Seq method has led to a wide range of discoveries. In plant and animal sciences, Iso-Seq has enabled investigators to produce high-quality genome annotations and conduct transcript expression analyses for agriculturally important crops, endangered species, model organisms, and evolutionarily distinct species. In human genetics, Iso-Seq has enabled scientist to elucidate rare diseases, distinguish medically important pseudogenes, create better cell atlases, and identify neoantigens as potential cancer vaccine candidates.
Breaking the throughput barrier with Kinnex
While Iso-Seq has established its critical value in plant and animal as well as human genetics studies, its relatively lower throughput can prohibit large-scale, population-level RNA experiments that could reveal significant isoform-driven insights. A typical Iso-Seq library on one SMRT Cell 8M on the Sequel II system generates approximately 4 million reads, which is sufficient for isoform discovery and genome annotation, but difficult to scale to large numbers of samples.
The new Kinnex kit has now shattered this barrier.
At ASHG 2023, three Kinnex kits covering three key applications were released to the world. The Kinnex full-length RNA kit for bulk RNA sequencing now enables users to produce 15 million reads per SMRT Cell on the Sequel II and 40 million reads per SMRT Cell on the Revio system, respectively. This represents a 16-fold throughput increase in RNA sequencing on Revio when using Kinnex. Because the Revio system gives users the flexibility to run 4 SMRT Cells simultaneously, sequencing time is also greatly reduced.
To illustrate this dramatic shift, PacBio and a team of collaborators generated a WTC-11 cell line dataset using 11 Revio SMRT Cells, totaling roughly 600 million reads in just three days. To generate an equivalent amount of data prior to the launch of the Revio system and without the Kinnex kit, it would have required 150 SMRT Cells and 38 weeks of run time!
Most importantly, this massive throughput increase does not compromise RNA transcript length or associated abundances found in the original sample. Kinnex gives users the same research advantage provided by long, accurate, full-length transcript information — now with more than an order-of-magnitude more reads in dramatically less time.
Learn more about this breakthrough capability in the Kinnex full-length RNA application note.
Not all reads are created equal
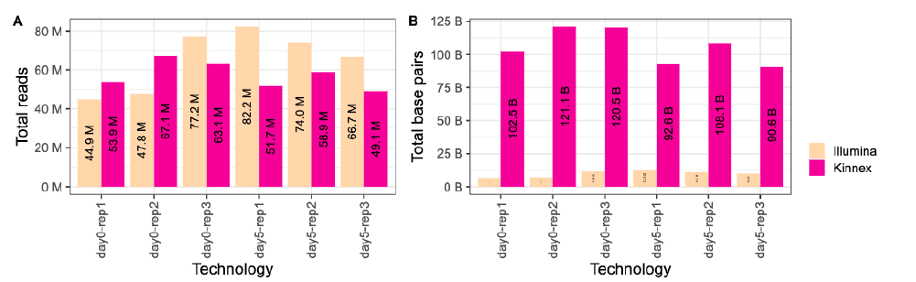
At the “read” level, the corresponding Kinnex and short-read data may appear to be comparable in terms of throughput. But at the “base pair” level, Kinnex offers users access to 10 times more data! This is because each Kinnex read represents a full-length cDNA molecule, which ranges in length from several hundred bases to over 10 kb. In contrast, each short read is only 150 bp long, offering only partial coverage of each full-length transcript. This means that short-read RNA-seq users must undertake the tedious task of computational transcript assembly, a process that has been repeatedly shown to lead to inaccurate estimates of transcript isoforms.
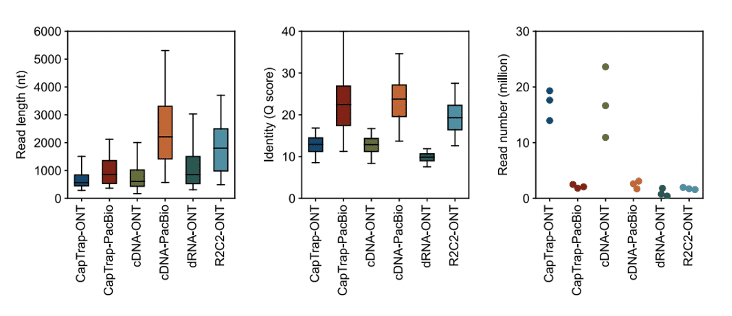
When compared with other long-read sequencing technologies, PacBio HiFi stands out for its accuracy. The LRGASP (Long-read RNA-seq Genome Annotation Assessment Project) consortium was an effort to systematically assess the performance of different long-read RNA-seq library preparation and analysis tools. Despite having 10 times fewer reads compared to Oxford Nanopore (ONT) data “on paper,” the consortium found that PacBio Iso-Seq data detected the greatest number of genes. The authors observed that “more reads did not consistently lead to more transcripts, indicating that read quality and length are important factors for transcript identification.” You can read more about the LRGASP study in this blog post or the bioRxiv preprint.
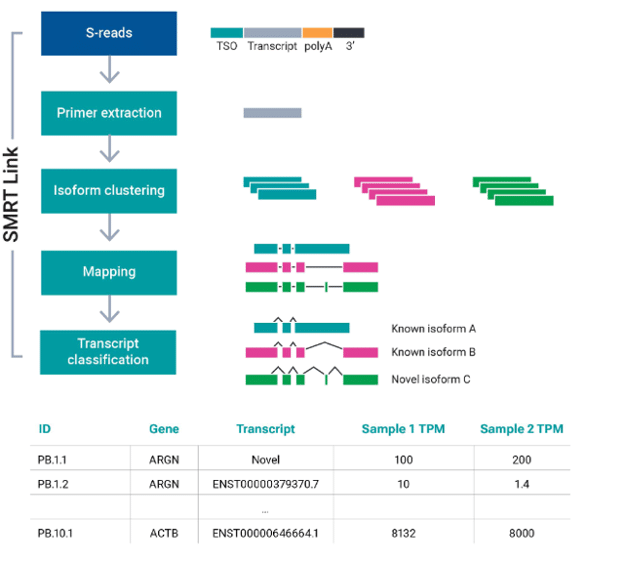
SMRT Link: Streamlined full-length transcript analysis by incorporating widely adopted community tools
New data types typically come with new computational challenges. In the early days of long-read RNA-seq, it was the analysis tools that users often found to be lacking. Solutions ported over from short read-based tools contained many residual assumptions about the fragmented nature of the data and could not take advantage of the fact that Iso-Seq data was full-length. Over the years, however, both PacBio scientists and the long-read user community have developed an ever-growing suite of dedicated long-read tools for transcript classification, genome annotation, quantitative analysis, fusion calling, and much more.
SMRT Link, the software used to run PacBio long-read sequencing instruments, now provides users with many built-in analysis capabilities. Starting with version 13.0, which was released at the same time as the new Kinnex kits, full-length transcript analysis is supported for bulk RNA sequencing in a streamlined push-button workflow. The “Read Segmentation and Iso-Seq“ analysis option in SMRT Link takes the HiFi reads generated from a Kinnex full-length RNA library and produces high-quality, full-length isoforms with classification and abundance information. At the heart of the transcript classification capability is pigeon, a PacBio adaptation of SQANTI3, which has become one of the most popular and accepted standards for classifying transcript isoforms against a known reference annotation.
SMRT Link 13.0 is just the beginning. For a curated list of more PacBio compatible tools for downstream RNA analysis, read our bioinformatics application brief.
Jumpstart tomorrow’s discoveries today
As the evolution of RNA sequencing continues, Kinnex emerges as a revolutionary force, enabling researchers to break through barriers and usher in a new era of possibilities. By delivering a more complete view of isoforms with unprecedented throughput, Kinnex exemplifies the commitment at PacBio to advancing transcriptomics. The recent launch of Kinnex kits enables HiFi sequencing users to generate 16-fold more reads without compromising data quality, transforming what is possible in RNA experiments.
The future of RNA-seq is now, and with Kinnex, researchers can get started on making the RNA discoveries of tomorrow, today. If you work in transcriptomics, now is the time to join the RNA revolution!
Download the Kinnex RNA datasets
Explore the Kinnex protocol and application note
Check out our webinars and events
Connect with a PacBio scientist