
In cities big and small, a hunt for an invisible threat unfolds right beneath our feet. Unseen but essential, our wastewater systems hold critical insights into public health.
Wastewater surveillance can be used to monitor new and recurring pathogens, their prevalence over time, and emerging variants – all providing valuable information on the development and spread of a disease throughout a community. While this practice gained widespread attention during the COVID-19 pandemic, it’s actually been a reliable tool for years, helping to track diseases like polio, hepatitis A, and influenza.
The recent advancement of culture-free methods, particularly through highly accurate metagenomic sequencing offered by the Onso system are opening doors to elevate this practice even further. With innovative sequencing tools, public health scientists can dig deep into genomic clues, serving as modern-day detectives in the fight against emerging health threats.
Read on to see how scientists are already witnessing the benefits of the new short-read sequencing SBB chemistry on Onso, enabling them to stay one step ahead in the ongoing battle to protect public health.
Understanding antimicrobial resistance through wastewater genomics
Beyond monitoring community diseases, Onso sequencing is being harnessed to tackle another pressing public health challenge: antimicrobial resistance. Antimicrobial resistance occurs when microbes like bacteria and fungi develop the ability to evade antimicrobials, including life-saving antibiotic medication. The World Health Organization has named the rise in antibiotic resistance as a top threat to global public health and development1, contributing to the deaths of at least 1.27 million people worldwide2.
In efforts to counter this urgent threat, scientists have identified antimicrobial resistance genes that allow bacteria to escape the effects of certain classes of antibiotics. Comprehensive examination of these genes requires highly accurate sequencing technology to properly identify ARGs and the types of antibiotics that they circumvent.
Recognizing this critical need, Zymo Research and PacBio have joined forces in a recent collaboration presented at this year’s ASM meeting. This partnership highlights the Onso system’s ability to deliver exceptionally accurate short-read sequencing at the resolution required to assess and combat antimicrobial resistance effectively.
PacBio and Zymo team up against antimicrobial resistance
The study that showcased this collaboration used Zymo’s Quick-DNA/RNA Water Kit to process real-world wastewater samples, sequencing them for a head-to-head comparison between the PacBio Onso system and the Illumina NextSeq 2000.
Between the two sequencing systems, the Onso platform generated more reads corresponding to resistance against a wide range of antibiotic class.
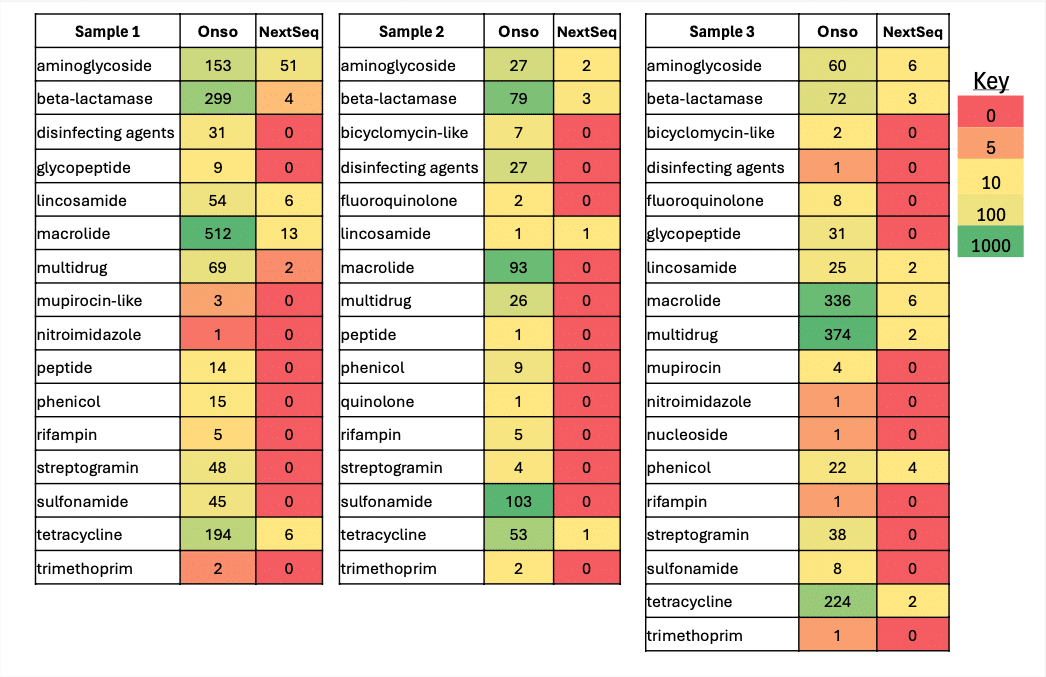
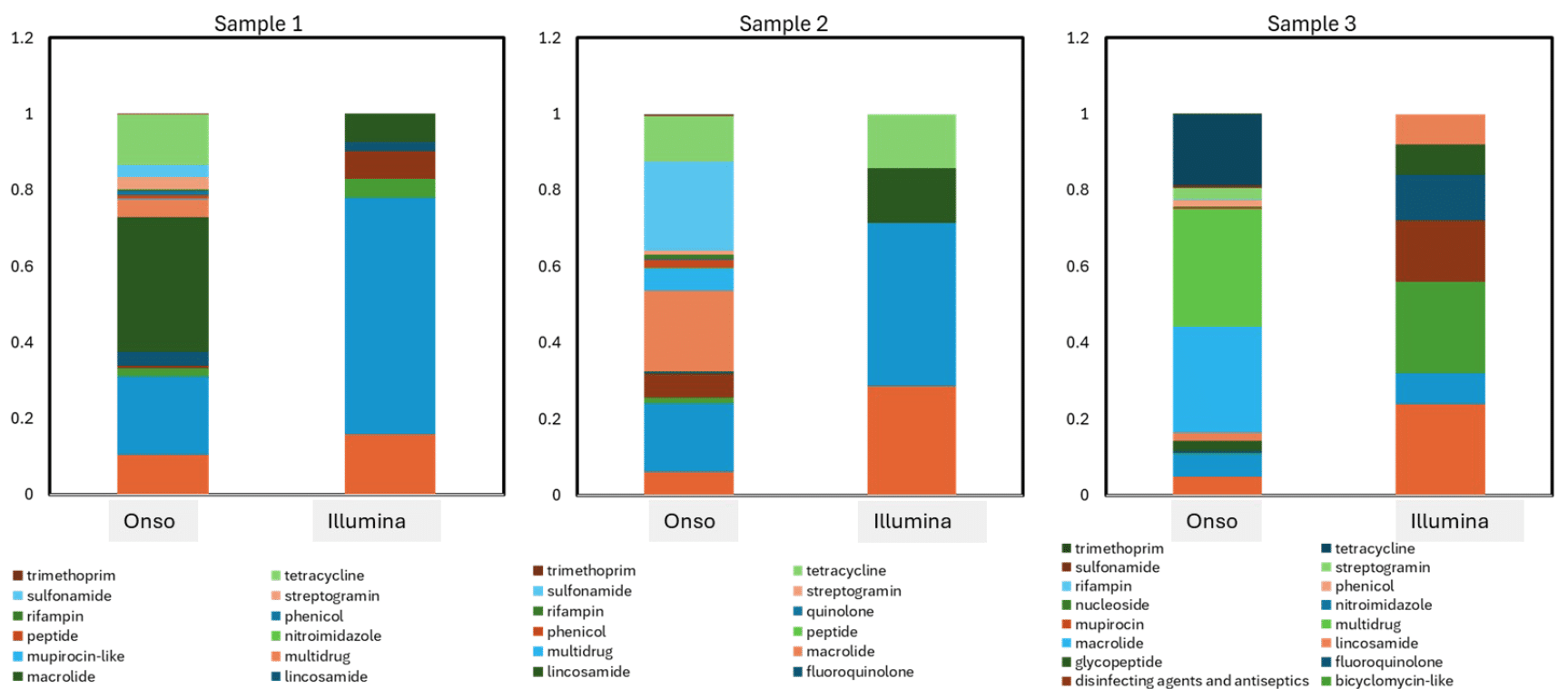
When assessed by relative abundance, this greater diversity revealed by the Onso system becomes all the more apparent. Whereas Illumina technology can detect only a handful of AMR classes in each sample, the PacBio Onso system displays a wider array of antibiotics under threat.
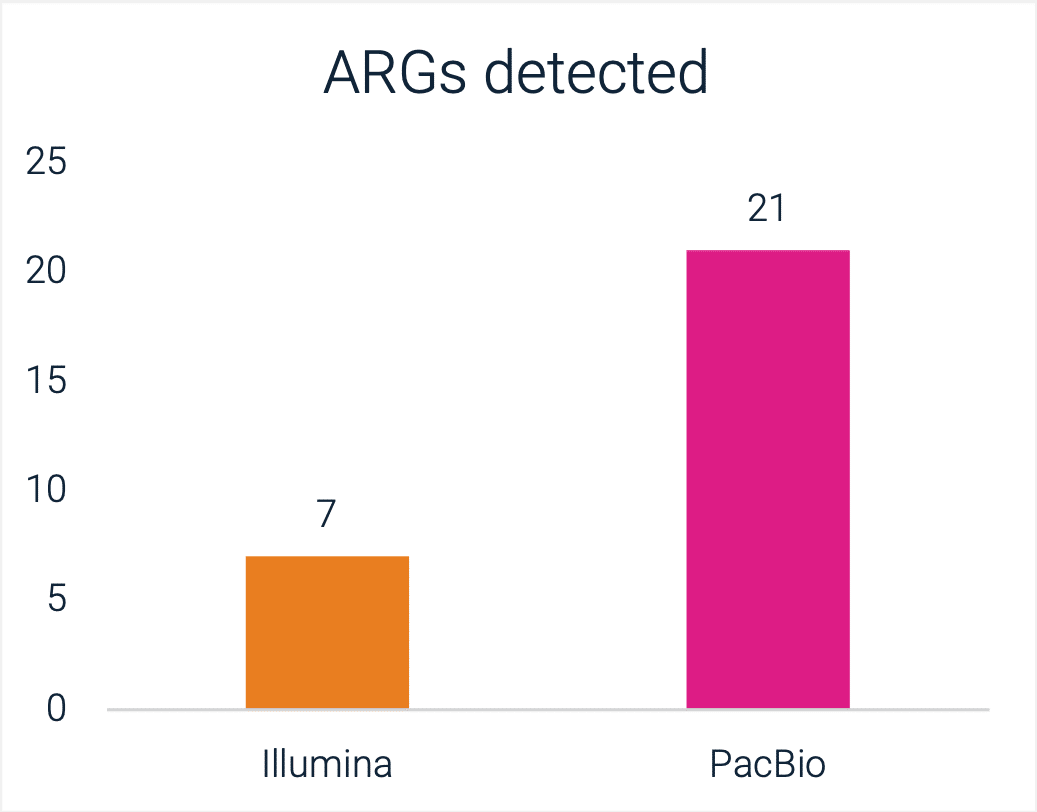
This is evidenced by the greater number of ARGs detected by the Onso system than NextSeq. Because many ARGs occur in pathogenic bacteria that can be transmitted to and among humans, these findings have the potential to directly inform public health decisions.
The conclusions of this study are a clear demonstration of the Onso system’s superior ability to detect infectious disease threats to public health. By providing exceptional accuracy in identifying antimicrobial resistance genes, the Onso system empowers researchers with the tools needed to anticipate and mitigate potential outbreaks:
“This precision equips researchers with the tools necessary for predicting disease outbreaks and optimizing both public health management strategies and water treatment processes, thereby bolstering environmental and public health outcomes.” – Cheng et al., 2024
At PacBio, we are invigorated by the transformative potential of our high-accuracy short-read sequencing technology. The ability to leverage such advanced tools in wastewater surveillance represents a pivotal step forward in safeguarding public health. We are committed to continuing this vital work, confident that the insights gained will pave the way for more proactive and effective public health interventions in the future.
Explore the benefits of Onso short-read sequencing for your research today
For a limited-time, PacBio is offering a discount designed to help you experience the cutting-edge SBB chemistry of the Onso system at a value that won’t be around for long.
References
- 1https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance
- 2https://www.cdc.gov/antimicrobial-resistance/about/index.html