Uncover the mysteries of RNA dysregulation in cancer
Our understanding of the complex biology of cancers is currently limited by the sequencing technology that we have available. For example, a study previously published in Science Advances showed that short-read based RNA-seq approaches only capture a faction of RNA isoforms that occur in breast cancer samples. On the flip side, long-read sequencing using PacBio Iso-Seq method detects ~2.5 times the number of isoforms, including a substantial number of novel isoforms. These aberrant transcripts are often translated into cancer-specific proteins and have been shown to affect cancer initiation, progression, metastasis, and drug resistance. As a result, accurate long-read sequencing can offer new potential clues into how to detect, track and treat various forms of cancer.
Long reads: more data, accuracy, and reliability
Until recently, many studies used a combination of short- and long-read technologies for isoform analyses. This was because long-read sequencing platforms lacked the necessary throughput for RNA quantification. But a new approach using PacBio HiFi technology has changed this.
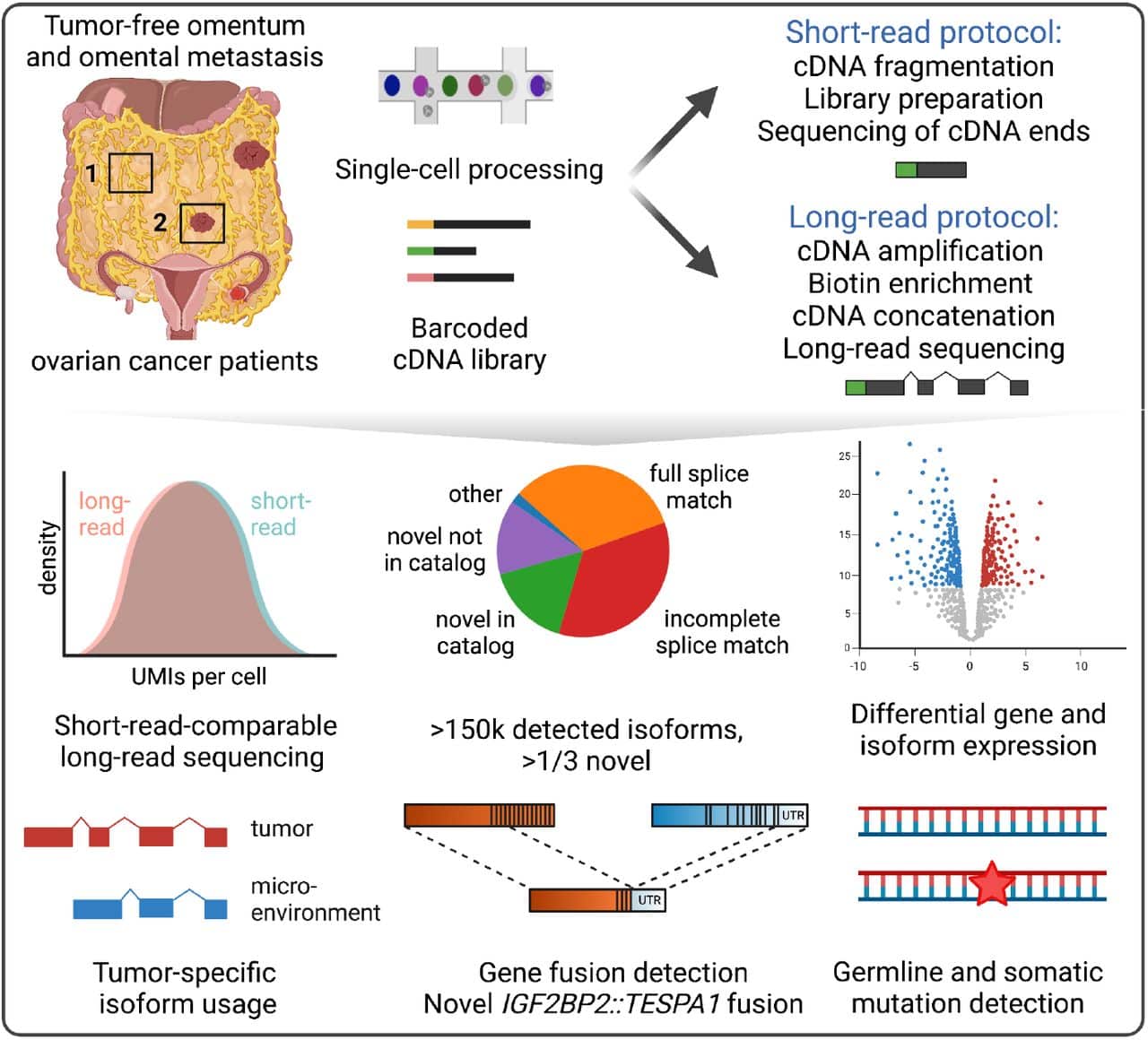
The Tumor Profiler Consortium took this new approach, using a now commercially available concatenation method to increase PacBio sequencing depth to be equivalent to that used for short-read experiments. This allowed them to perform cell type clustering and cell population identification without the use of short-read sequencing. Their findings indicate that long-read scRNA-seq is not a complementary technology, but rather a replacement for traditional short-read sequencing.
 This study highlights the benefits of accurate long-read RNA sequencing for RNA dysregulation in cancer, beyond gene expression. By leveraging a novel high throughput, single-cell method, the authors were able to detect various RNA “variants” in tumor samples, including RNA fusions, isoforms, and expressed mutations.
This study highlights the benefits of accurate long-read RNA sequencing for RNA dysregulation in cancer, beyond gene expression. By leveraging a novel high throughput, single-cell method, the authors were able to detect various RNA “variants” in tumor samples, including RNA fusions, isoforms, and expressed mutations.
Using the largest PacBio single-cell RNA sequencing dataset to date, the study revealed novel mechanisms associated with cell transitions in ovarian cancer-associated cell populations. Further analyses also revealed novel fusions that were misclassified as abberant gene expression changes in matched short-read data.
The evidence is clear – the time has arrived where researchers can now use long-read sequencing to confidently identify cell types without the help of short-read sequencing. This is a huge breakthrough in cancer research, as it allows researchers to study cancer at a much deeper level – achieving everything that traditional short-read technology can, and more.
With PacBio long-read sequencing, cancer researchers can now:
1. Achieve gene and isoform expression analyses
2. Discover novel isoforms
3. Detect known and novel fusions
4. Accurately detect cancer-related expressed mutations
With the findings of this study, the authors of the paper conclude that:
“Long-read sequencing provides a more complete picture of cancer-specific changes. These findings highlight the manifold advantages and new opportunities that this technology provides to the field of precision oncology, opening the premise of personalized drug prediction and neoantigen detection for cancer vaccines.”
You can learn more about uncovering cancer-specific RNA isoforms using long-read sequencing in our application brief. You may also connect with a PacBio scientist to learn more about HiFi sequencing and solutions for cancer research.