
Microbiology forms the bedrock of modern genetics and biotechnology. Whether its basic science, alternative energy development, drug discovery, or human health – our planet’s smallest organisms play a critical role in the bigger picture. Even so, most microbial life remains unknown to science. And that’s why cutting-edge microbial ecology and metagenomics research seeks to reveal the biological secrets of this hidden majority. But how do we go about understanding what that we have, thus far, struggled to observe in detail? The answer? Sequencing. Really good sequencing.
Read on to discover:
- Why HiFi sequencing is the best choice for microbiome and metagenomics research.
- What makes the new Kinnex 16S rRNA kit and Revio system a game-changer for microbiology.
- Details on new downloadable HiFi microbiome and metagenomics datasets.
I’ll read the details later. Take me to the datasets!
HiFi sequencing for breakthrough microbiome discoveries
HiFi reads generated by PacBio long-read sequencing systems enable microbiologists to access metagenomic data with entirely new levels of information density and discovery potential. By leveraging reads with an average length of up to 20 kb and an accuracy of >Q30, PacBio HiFi users have access to the contiguity and completeness necessary to generate reference quality metagenome assembled genomes (MAGs). This is true for even very low-abundance taxa. At the same time, HiFi shotgun metagenome sequencing yields stunningly precise taxonomic profiles while simultaneously generating richer and more abundant functional information than short-read sequencing alternatives. HiFi sequencing users can also take advantage of long reads to analyze full-length 16S rRNA amplicons at high taxonomic resolution that would otherwise require shotgun metagenomics data to obtain.
Kinnex 16S + Revio take the best in microbiome sequencing and make it better
More data, more value, more insights.
Building on the trusted performance of the Sequel II generation, PacBio has changed the game again in microbial ecology and metagenomics with the release of the Revio system. The massive 15x throughput increase on this powerful platform enables microbiologists to tackle larger and more complex samples, sequence deeper, and batch samples more efficiently all while delivering extraordinary data quality.
And now, in the fall of 2023, PacBio is pleased to present the Kinnex 16S rRNA kit for PacBio long-read sequencing platforms. This exciting new kitted solution enables microbiologists using Sequel II and Sequel IIe and Revio systems to boost 16S taxonomic data generation by nearly an order of magnitude! The Kinnex 16S rRNA kit is a versatile throughput boosting solution based on the MAS-Seq method developed by scientists at the Broad Institute of MIT and Harvard. Kinnex 16S rRNA works by taking advantage of very long HiFi read lengths to concatenate multiple full-length 16S amplicons into single molecules which yields 8 to 12-fold more 16S reads per sample. This big bump in throughput makes full-length 16S HiFi sequencing scalable and more affordable while providing extraordinary taxonomic resolution.
Ready for download: new HiFi metagenomics and Kinnex 16S datasets
Over the last couple of years PacBio has worked closely with Zymo Research –a leading developer of biomedical research products– to characterize the ZymoBIOMICS Fecal Reference with TruMatrix Technology. This rigorously developed sample is a leading reference standard for use in benchmarking and controlled studies of the human gut microbiome. All the datasets shared (except the mock communities) in this release analyze this microbiological reference.
The datasets
HiFi full-length 16S rRNA gene using Kinnex 16S data
- Dataset 1: ZymoBIOMICS Fecal Reference with TruMatrix Technology (Revio and Sequel IIe, Kinnex 16S and monomer 16S). See Table 1.
- Dataset 2: Mock community microbiome standards (Revio and Sequel IIe, Kinnex 16S).
HiFi shotgun metagenomics data
- Dataset 3: ZymoBIOMICS Fecal Reference with TruMatrix Technology on Revio. See Table 2.
Background on the new microbiome datasets and the PacBio analysis approach
How we analyzed the datasets
The collaboration between PacBio and Zymo Research focused on metagenome assembly and MAG recovery with a combined dataset and merged MAG approach. The sequencing was performed on both the Revio and Sequel IIe platforms. HiFi reads were then assembled with hifiasm-meta and binning of the assembled contigs was performed using two approaches, the HiFi-MAG-Pipeline along with Phase Genomics’ ProxiMeta (Figure 1). Kinnex 16S rRNA data was analyzed using the PacBio pb-16S-nf pipeline.
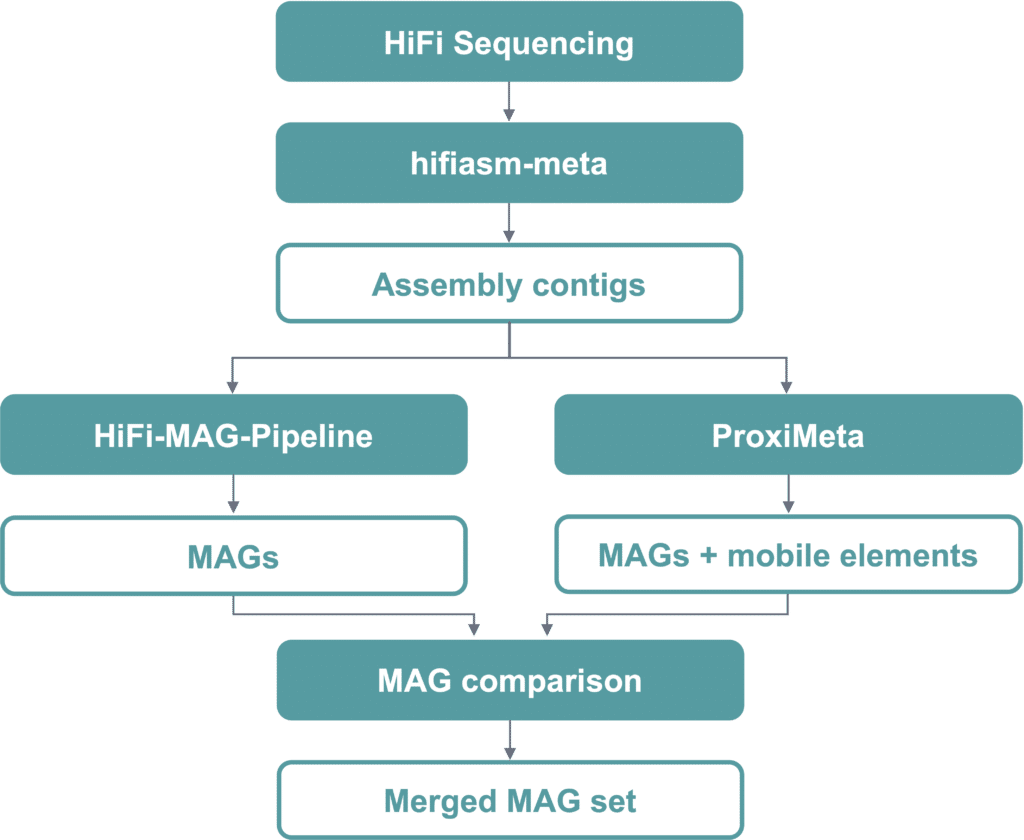
Figure 1. The merged bioinformatics workflow used to generate metagenome assembled genome datasets from the ZymoBIOMICS Fecal Reference with TruMatrix Technology.
Examples of our analysis performance
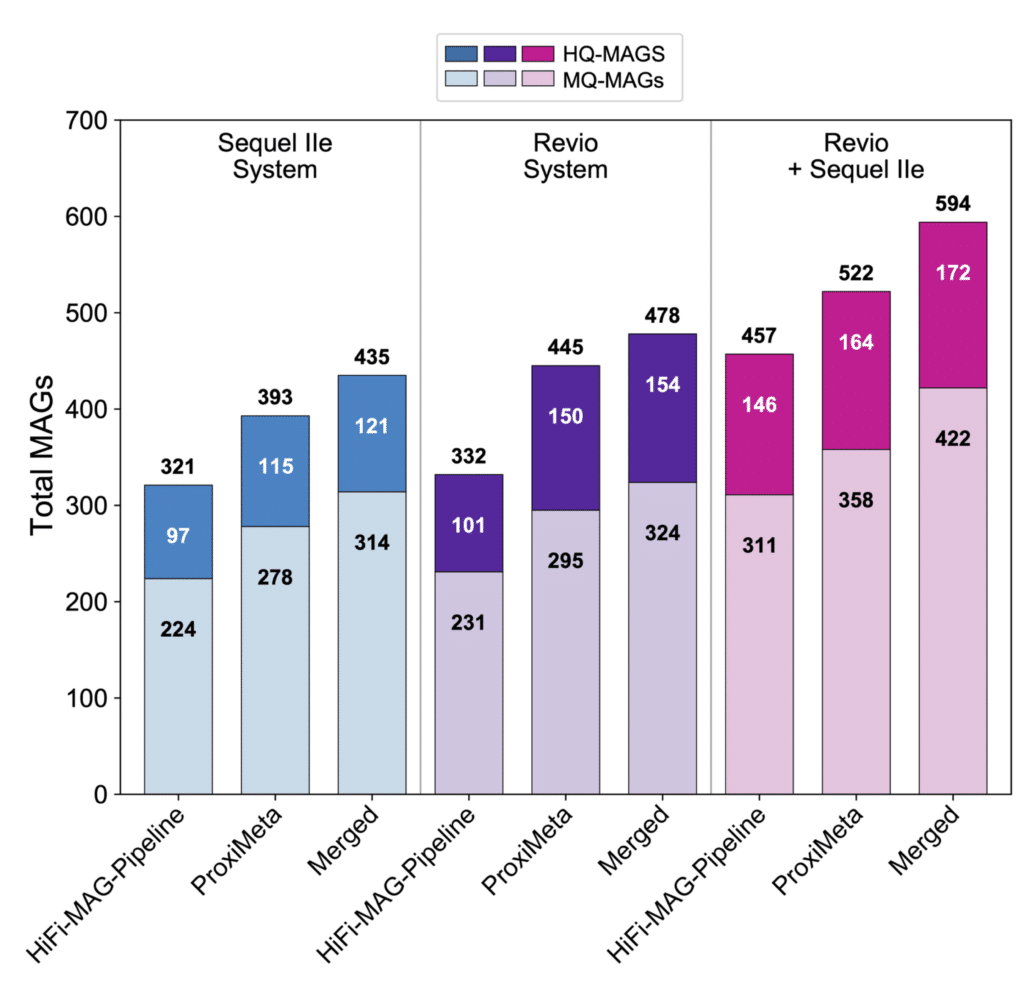
Figure 2. Total number of high-quality MAGS (HQ-MAGs) and medium-quality MAGS (MQ-MAGs) assembled during an analysis of the ZymoBIOMICS Fecal Reference using PacBio HiFi metagenome sequencing. MAGs were identified using the HiFi-MAG-Pipeline, Proximeta, and a merged approach as outlined in figure 1. Separate studies of the reference sample were performed on the Sequel IIe and Revio systems. Findings from these two separate experiments were combined to create a catalog of total unique MAGs discovered within this Zymo reference sample.
Table 1. Kinnex 16S performance metrics for ZymoBIOMICS Fecal Reference with TruMatrix Technology analyzed on Sequel IIe and Revio platforms.
| Dataset | Input HiFi reads | Segmented reads (S-reads) | Mean length of S-reads | Percentage of reads with full array | Median QV |
| Sequel IIe System Zymo Fecal Reference Kinnex 16S | 2.55M | 30.47M | 1551 | 96.75 | Q33 |
| Revio System Zymo Fecal Reference Kinnex 16S | 4.65M | 55.26M | 1551 | 94.70 | Q30 |
Table 2. Shotgun metagenome data metrics for ZymoBIOMICS Fecal Reference with TruMatrix Technology
| Datasets | HiFi yield | HiFi reads | Average HiFi insert length | Median QV |
| Sequel IIe system 6 SMRT Cells (8M) | 120.8 Gb | 17.1 M | 7.0 kb | Q40 |
| Revio system 2 SMRT Cells (25M) | 135.1 Gb | 17.6 M | 7.4 kb | Q46 |
| Combined | 255.9 Gb | 34.7 M | 7.7 kb | Q42 |
- Download PacBio HiFi datasets and see the HiFi difference for yourself!
- Check out the Zymo Research fecal reference database
Welcome to the future of metagenomics
From time-to-time new methods and new technological capabilities are needed to help drive the frontier of knowledge forward. For microbial ecology and metagenomics, systems like Revio and advanced kits like Kinnex 16S can enable you to do just that. Already there have been 50+ publications and pre-prints to-date utilizing PacBio HiFi for shotgun metagenomics with many more on microbiome amplicon sequencing (16S / ITS / 18S / etc.). With this growing body of research and these publicly available datasets, we believe you’ll see that HiFi sequencing provides a fresh approach to microbiome research. Tackle today’s biggest questions in microbiology with tomorrow’s metagenomic technology.
Connect with a PacBio scientist to join the future of metagenomics