
Highly accurate long-read sequencing technologies have made it possible to generate genome assemblies at scale. In fact, HiFi sequencing is now the technology of choice for assembling reference-quality genomes due to its unique combination of length and 99.9% accuracy, resulting in more contiguous and complete assemblies. The latest SPRQ chemistry update on the Revio system enables long-read sequencing from as little as 500 ng of DNA, marking a pivotal moment for researchers studying complex genomes.
But what if you need to sequence ultra-limited samples or explore the genomes of small organisms that have been historically difficult to decode? Enter the Ampli-Fi protocol—a new workflow that will enable HiFi sequencing from as little as 1 ng of genomic DNA.
Curious about how this protocol will reshape the sequencing landscape for underrepresented species? Keep reading to see it in action and discover what’s next.
As little as 1 ng DNA input for all HiFi sequencing systems
A few years ago, PacBio released low-input and ultra-low-input protocols for HiFi sequencing. At the time, the definition of “ultra low” was 5 ng—enough to enable scientists to sequence tiny arthropods. But we didn’t stop there. Times have changed, and the need for even lower inputs has persisted.
The new Ampli-Fi workflow represents the latest evolution in PacBio’s low-input solutions for scientists studying the world around us, from the tiniest species on the tree of life to preserved specimens from ancient eras of history. Becoming available by the end of Q1 this year, the Ampli-Fi workflow replaces the previous “ultra-low-input” protocol, enabling sequencing from as little as 1 ng of DNA for all HiFi sequencing systems, including Revio and the new Vega benchtop system.
In addition to requiring only a fraction of the previous DNA input amounts, the Ampli-Fi protocol improves upon the older protocols by supporting species with larger genomes (up to 3 Gb) and reducing library prep costs, thanks to the SMRTbell prep kit 3.0.
| Ampli-Fi protocol | Ultra-low protocol with gDNA amplification kit |
|---|---|
| Available on Vega, Revio, and Sequel II systems | Only available on Sequel II systems |
| Single amplification reaction | Two separate amplification reactions |
| 1 – 50 ng DNA input | 5 – 20 ng DNA input |
| Supports genomes up to 3 Gb | Supports genomes up to 500 Mb |
| Compatible with SMRTbell prep kit 3.0 | Required older library prep kits |
With the new Ampli-Fi protocol, you can now sequence previously inaccessible samples, such as certain types of metagenomes, individual insects or small organisms, archival specimens, dried blood spots, and tumor tissue using HiFi sequencing on the Vega and Revio systems.
Better genome assemblies for difficult-to-sequence species
What do insects, the sea slug, and other tiny critters all have in common? They were all notoriously difficult to sequence using long-read technology… until now.
The new Ampli-Fi protocol uses the KOD Xtreme™ Hot Start DNA polymerase to amplify 8-10 kb DNA fragments that have been ligated with a universal PCR adapter. This particular polymerase is known to reduce PCR bias, especially in high-GC regions, and generate more contiguous genome assemblies compared to others.
In a new preprint titled Long-read sequencing and genome assembly of natural history collection samples and challenging specimens, Bein B, et al. (2025) use the KOD Xtreme Hot Start DNA polymerase to generate a high-quality genome assembly of the maned sloth, Bradypus torquatus.1 They demonstrate that the KOD XTreme Hot Start polymerase (labeled polymerase C) reduces bias and results in a more contiguous genome assembly compared to alternate polymerases A and B (Figure 1). In contrast, polymerases A and B resulted in regions of the genome with no coverage, creating gaps in the assembly.
Figure 1: Contiguity of B. torquatus assemblies generated with data from ultra-low input libraries prepared with polymerases A/B and/or C at different coverages
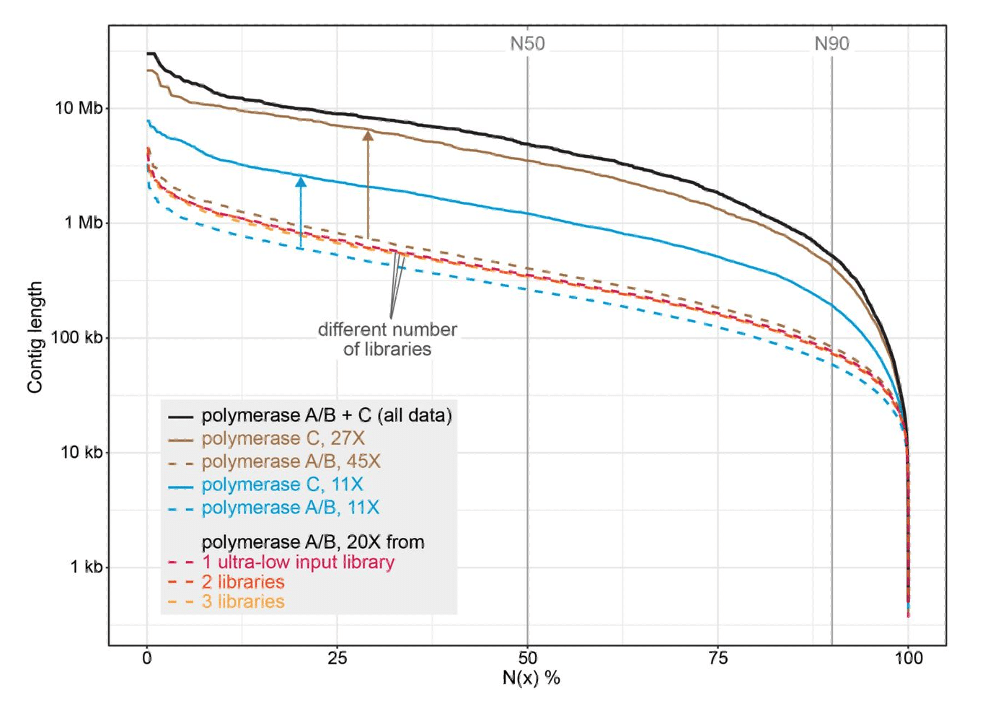
The researchers applied this protocol to other ethanol-preserved museum specimens, showing that it also improves the contiguity of assemblies of several mollusk and arthropod species.
“The efficacy of this protocol facilitates genome assembly of challenging taxa and suggests that collections can serve as valuable sample sources for long-read sequencing.”
—Bein B, et al. (2025)
In another study, (Männer et al., 2024) use the KOD Xtreme Hot Start DNA polymerase protocol to generate a chromosome-level genome assembly and annotation of the marine sacoglossan sea slug, Elysia timida.2 This assembly resulted in nearly 20,000 protein-coding genes, which are more contiguous and complete compared to other publicly available annotations of Sacoglossa.
“…Our methodological approach helps to obtain a high-quality genome assembly even for a “difficult-to-sequence” organism.”
—Männer L, et al. (2024)
In both examples, the authors show that the Ampli-Fi protocol can be used to overcome the challenges of sequencing DNA containing damage, contaminants, or other sequence inhibitors. These types of samples are incompatible with amplification-free methods. Using the Ampli-Fi protocol, researchers can generate high-quality assemblies from even these difficult sample types.
Small input, big impact: Ampli-Fi opens new doors for diverse species
The new Ampli-Fi protocol will be available by March 31, 2025. Contact us to be the first to hear about this new protocol and other innovations for plant and animal genomics.
References
- Bein B, Chrysostomakis I, Arantes LS, et al. (2025) Long-read sequencing and genome assembly of natural history collection samples and challenging specimens Genome Biology 26:25.
- Männer L, Schell T, Spies J, et al. Chromosome-level genome assembly of the sacoglossan sea slug Elysia timida (Risso, 1818). bioRxiv 2024.06.04.597355; doi: https://doi.org/10.1101/2024.06.04.597355