
The new Revio long-read sequencing system, with its first customer shipment in March 2023, has changed the game in genomics. It enables researchers to access an entirely new echelon of discovery opportunity. And while the system’s applications and advantages in the human space are substantial and rightly touted, the potential of the Revio system to enable a revolution in plant and animal genomics might be even more significant.
Change what you expect from plant and animal genome sequencing technology
During our formative years of scientific training, the methods that predominate can shape the scientific questions we ask and influence our perception of the natural world. Many of today’s genomics experts began their careers during eras of DNA sequencing technology that were led by chain-termination sequencing and later, high-throughput short reads pioneered by Solexa and Illumina.
Nowadays, the technological landscape that underpins genomic research looks quite different than it did even five years ago. PacBio long-read sequencing is a highly accurate, scalable technology with impressive throughput that compels the scientific community to fundamentally reconsider what is achievable and break free from outdated practices. The Revio long-read sequencing system enables researchers to generate 360 Gb of long-read data every 24 hours with reads averaging 15-20 kb in length at Q30+ accuracy, and native 5mC epigenetic detection in CpG contexts included.
In the age of Revio, genomic complexity just got a whole lot easier
The greatly expanded capacity to generate PacBio long reads offered by the Revio system enables plant and animal researchers to confidently surmount the technical roadblocks of old to make more impactful discoveries. With this new platform researchers can now make phased de novo assemblies of everything from relatively simple single-celled organisms like S. cerevisiae to the titanic 90 Gb genome of the European mistletoe –often faster and more easily than you would think.
In a shining example of what’s now possible with the Revio system, Katherine Jenike, a PhD candidate in Mike Schatz’s group at John’s Hopkins University leveraged the throughput of the platform to accomplish an impressive feat of sequencing. Jenike was able to sequence 8 members of the nightshade family in 24 hours using 4 Revio SMRT Cells. She then took the resulting 360 Gb of HiFi long-read data from these samples and constructed a pangenome of the group using Panagram, a tool she developed at Johns Hopkins. Accomplishments like this are exciting because they underscore what the latest PacBio long-read technology can enable: pangenomes in a day.
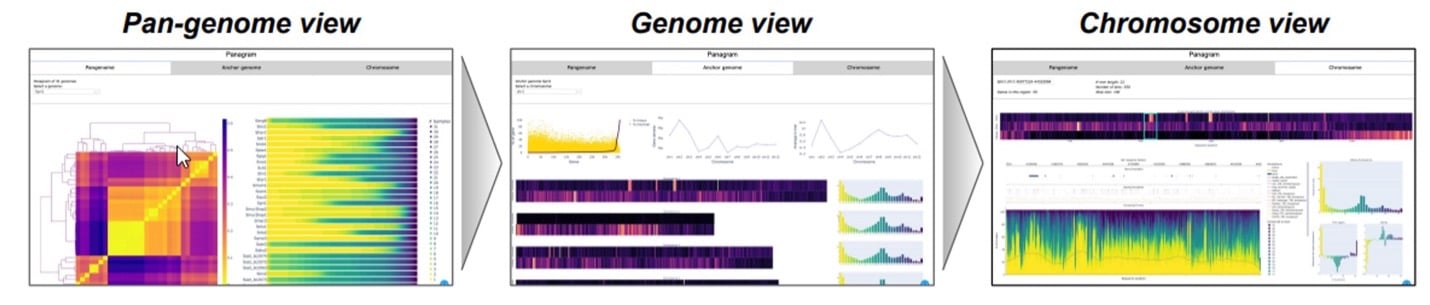
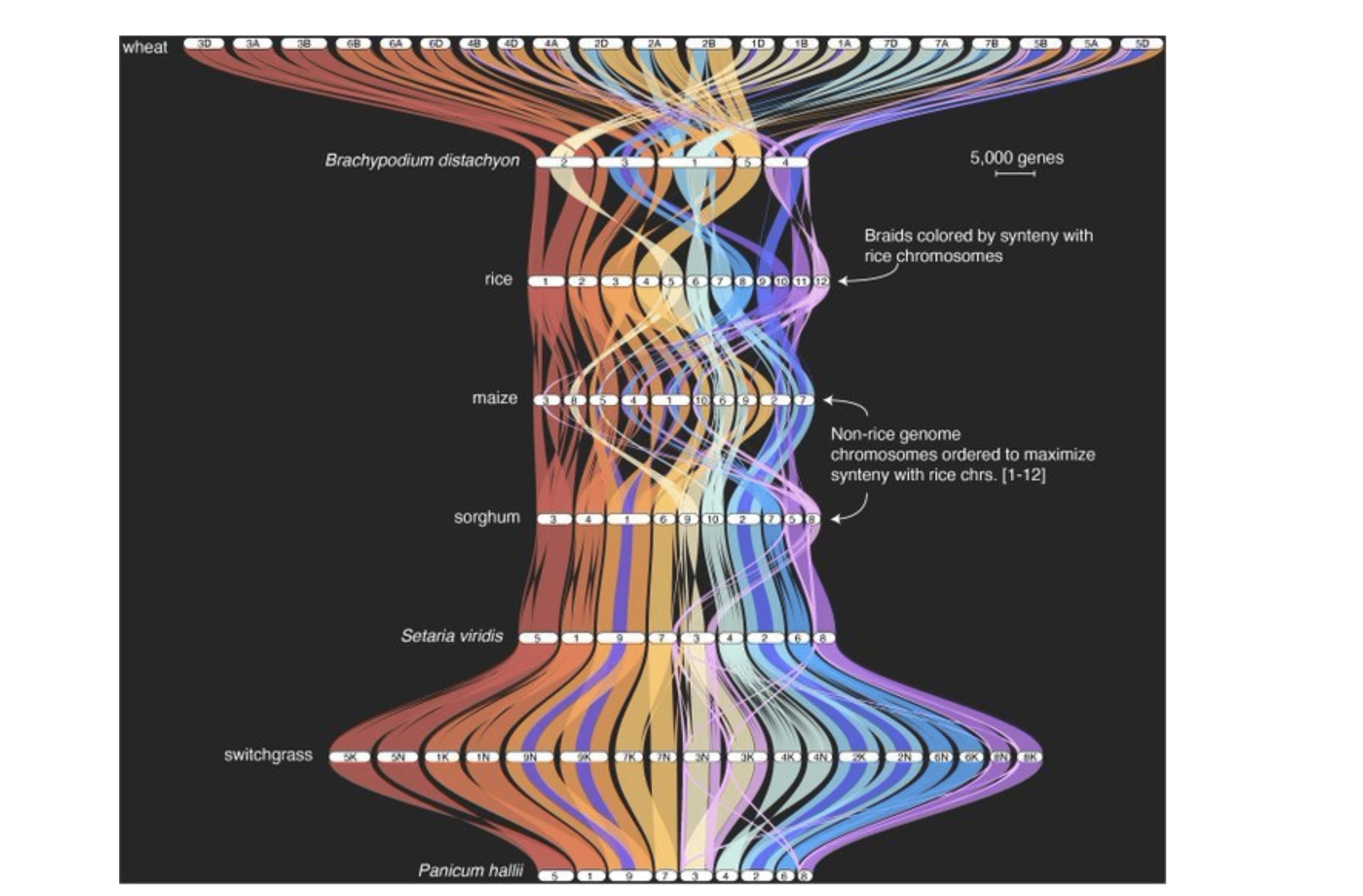
In response to this newfound accessibility to the power of pangenome analysis, the growth of accompanying ecosystem of analysis tools is also accelerating. In addition to Panagram, John Lovell’s GENESPACE is another tool that is available to help accelerate discovery using pangenomes with impressive visualization features. So, whether you study maize, salamanders, or human diseases: in the age of Revio, genomic complexity just got a whole lot easier.
Speed and efficiency to drive innovation on a grand scale
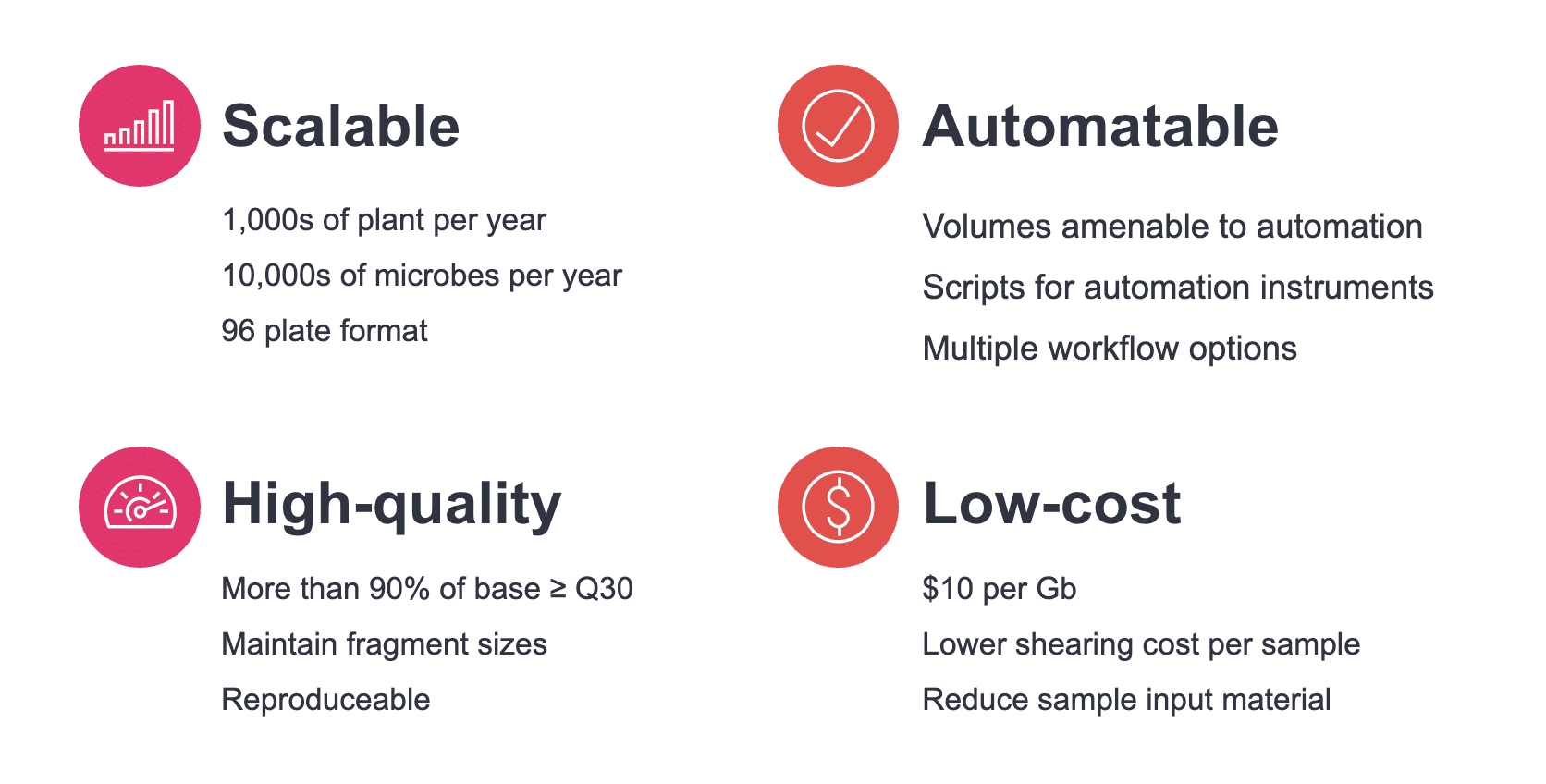
The Revio system has been so successful at making high-quality long reads more accessible that the constraints on industrial-scale long-read genomics have begun to shift from the sequencing stage to the broader sample preparation workflow for some users.
In response to this challenge, PacBio and industry collaborators recently released high-throughput workflow protocols for sequencing plant and microbial genomes on a massive scale using the new Revio system. This protocol is now publicly available and can enable users to sequence thousands of plant genomes and tens of thousands of microbial genomes per year. Dramatic increases in sequencing capability like this can greatly enhance the global push to optimize our food systems in ways that boost both productivity and nutrition while potentially enhancing sustainable land-use practices.
Changing the game and changing the name
Paired with the throughput of the Revio system, the length and high accuracy of HiFi long-read data now make it possible for researchers to confidently sequence through regions of high homology to identify long-ranging structural variants that are practically invisible to short-read technologies. In this way the Revio system and PacBio long reads are changing the game right down to the lexicon used by researchers to discuss the genomes they study. Regions deemed to be “dark”, “highly complex”, or “hard to sequence” are rooted in the limitations of the short-read instruments that defined previous technological eras of discovery. With contemporary PacBio sequencing technology, genomic regions that fit this description are dwindling and may go away altogether in the future for some organisms. As a result, what was once in the dark is now out in the light; the complex is becoming straightforward; and the hard-to-sequence has become simple.
Making high-quality big data more accessible for individual labs
The diversity of genomic configurations found across the tree of life is staggering. When we look beyond humans, genome structure, size, and ploidy expand considerably. To make sense of the diversity, researchers have often had to conduct vast amounts of sequencing on subject organisms –a task that is both time consuming and expensive. The result? A de facto limit on the type of research questions that can be pursued by more modestly funded research teams. Luckily, HiFi long-read sequencing and the launch of the Revio system is changing that.
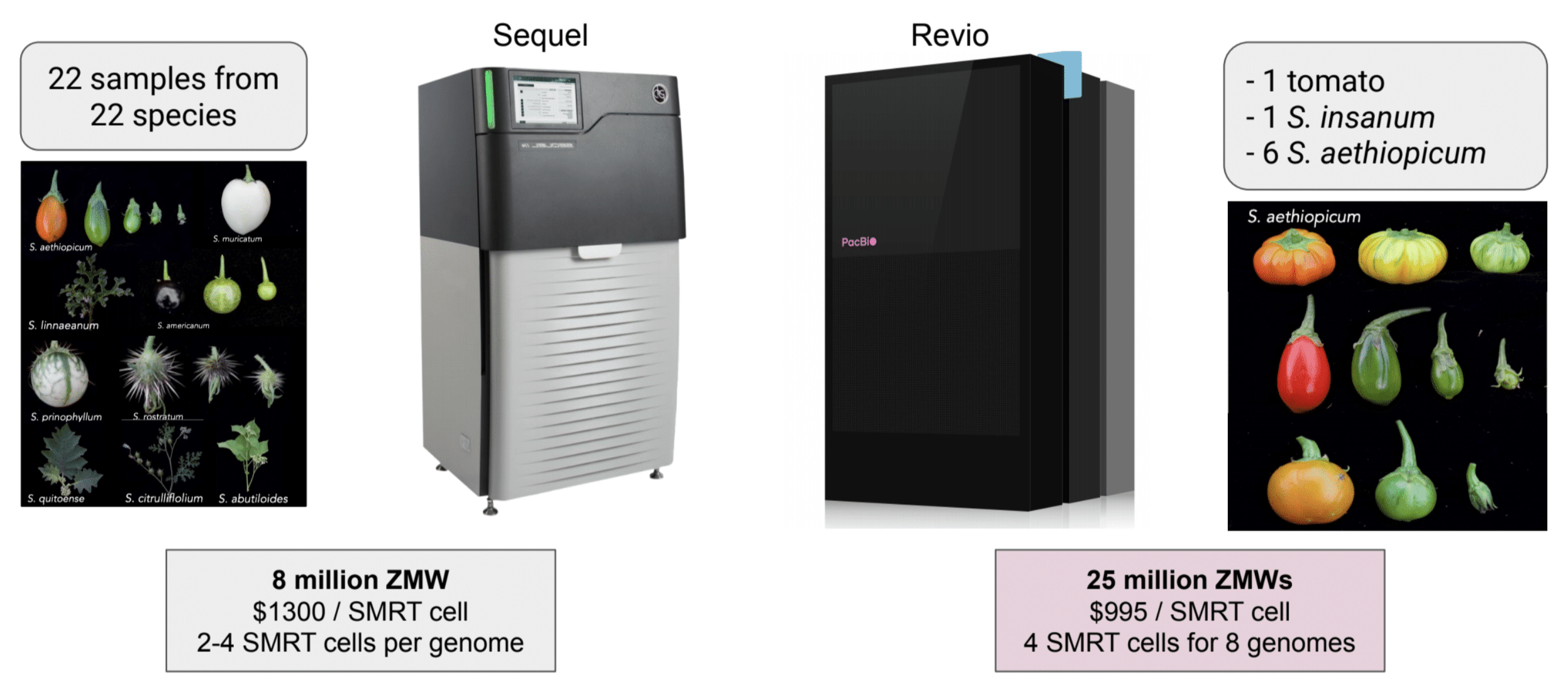
Returning to our example of nightshade genomics, scientists in the Schatz lab at Johns Hopkins conducted much of their previous work on this family of plants using an earlier generation of PacBio long-read sequencers. The team sequenced 22 species using 22 samples for cost of around USD $2800 per sample*. In addition to this price tag, each nightshade genome required about 60 hours to sequence at the time. Fast forward to today and the same research project would now cost almost six times less with the launch of the latest PacBio long-read system. On a Revio instrument, each nightshade genome would now cost approximately USD $497.50 (when batched) with a turnaround time of only 24 hours. Such significant drops in cost like this mean more researchers than ever will be empowered to pursue bigger questions in plant and animal genomics with more comprehensive high-quality data in hand.
With the Revio system and a growing ecosystem of analysis tools, research teams and laboratories great and small can now generate big data to drive high impact discoveries in plant and animal genomics.
Rethink what is possible. Are you ready for Revio?
Explore how you can access the big data needed to tackle complex genomes at scale and speak with a PacBio scientist to see how to get started.
Check out these additional resources:
Plant + animal genomics with PacBio
Plant + animal HT workflow protocols
PacBio certified sequencing service providers