Most higher organisms are diploid, meaning that each cell carries in its nucleus two copies of the individual’s genome, one from its mother and one from its father. Accurately separating and assembling these two copies, or haplotypes, has challenged the genomics community because the two copies are very similar to each other (they are from the same species), but not identical – if they were, your mother and father would look the same! Accurate and contiguous information about these small differences over extended genomic regions is needed to separate, or phase, the two haplotypes. Because short-read sequencing cannot provide such information, in the past genome assemblies have typically been expressed as a single sequence – representing a collapsed mixture of the two haplotypes – which is actually not present in the organism.
The performance of different long-read sequencing technologies to phase the two haplotypes in a genome assembly was recently compared by Duan et al. (2022). The researchers cleverly took advantage of the fact that many fungi have multiple haploid nuclei per cell, i.e., they neatly package one full set of haploid chromosomes into separate nuclei.
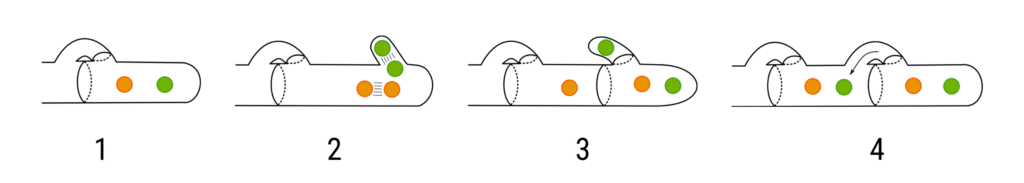
This physical separation allows for establishing the ground truth for the two haplotypes, which in turn permits benchmarking of the phasing accuracy from different sequencing technologies, here evaluated for PacBio HiFi sequencing and Oxford Nanopore Technologies (ONT).
The results were strikingly different. The table below summarizes the researchers’ findings:
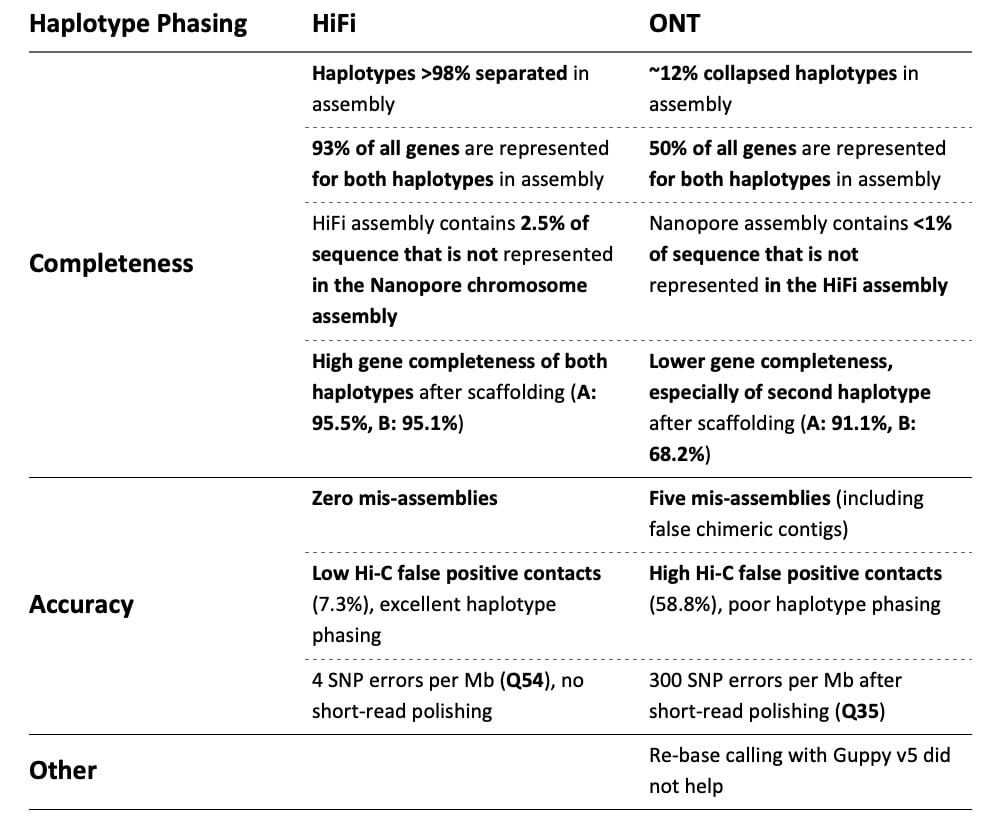
As the final result, the HiFi assembly was scaffolded with Hi-C data into a curated, reference-quality assembly that accurately and fully represents the diploid nature of this organism (below). In contrast, the authors noted that for ONT, “the presence of extensive phase switches in this assembly precludes the accurate separation of haplotypes.”
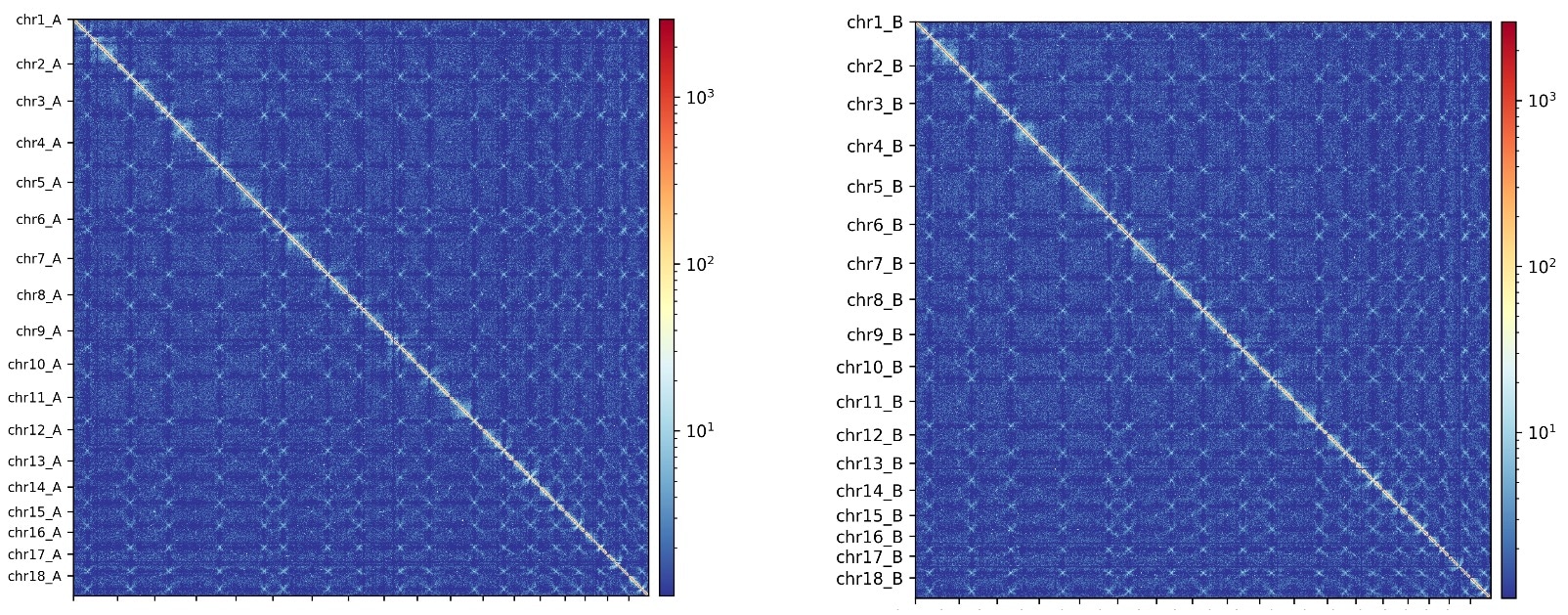
Similar findings have been reported in numerous other publications. For example, a recent preprint by the Human Pangenome Reference Consortium (HPRC), describing an extensive comparison of many different sequencing technologies and assembly methods for the automated assembly of high-quality diploid human reference genomes, observed that PacBio HiFi sequencing resulted in the best performance. And of course in the area of plant genomics, the haplotype phasing of even more challenging polyploid genomes is now addressable with HiFi sequencing, described for the tetraploid rose genome and the octoploid strawberry genome as just two examples.
For a long time, because of technological limitations, the genomics community had to settle for collapsed genome assemblies, thereby forgoing important biological insights, preventing discoveries, and hampering our understanding of the true complexity and workings of diploid genomes. Thanks to PacBio’s HiFi sequencing, providing the necessary combination of highly accurate and long sequence reads, fully phased diploid genome assemblies can now be routinely generated, allowing for the true genome representation of the biological sample, and revealing the full picture of an organism’s genomics.
Read more about the HiFi Difference
More precise genomes for precision medicine
Not all gigabases are created equal
True long reads vs. synthetic long reads
Looking for additional resources?
HiFi sequencing
Diploid genome assembly and comprehensive haplotype sequence reconstruction
Sequencing 101: From DNA to discovery – the steps of SMRT sequencing