
The combined accuracy, read length, and coverage uniformity of PacBio HiFi sequence reads enables the characterization of many previously inaccessible regions in the genome. Two new preprints by Oscar Rodriguez, William Gibson, Melissa Smith, Corey Watson (University of Louisville, KY) and collaborators describe the use of HiFi sequencing to illuminate immunoglobulins (say that fast three times!), providing another powerful demonstration of how much there is still to discover and learn about human genomes. The studies also demonstrate the use of targeted HiFi sequencing best practices to study regions of interest at low cost and with high throughput.
The first preprint, entitled Antibody repertoire gene usage is explained by common genetic variants in the immunoglobulin heavy chain locus characterizes the IGH region, one of the three genomic loci that constitute the adaptive immune system through encoding antibody genes. As the authors describe, its “extensive haplotype diversity and locus structural complexity has made IGH haplotype characterization challenging using standard high-throughput approaches, and as a result it has been largely ignored by genome-wide studies”, and that “microarray and short-read sequencing are not able to fully and accurately resolve IG germline variation”. Now, using HiFi sequencing they comprehensively assembled the IGH loci and genotyped IGH variations in 154 healthy adults, providing “the first comprehensive picture of IGH polymorphism”. The extensive IGH variation was striking and included novel structural variants (SVs, collectively spanning over half a megabase), small indels and SNVs, and even entirely novel IGH genes and alleles. Traditionally, SNVs and indels are difficult to detect and genotype in segmental duplications and within SVs – here, in the haplotype-resolved HiFi assemblies cataloged “a total of 4,625 (23%) SNVs had not been previously identified cataloged in dbSNP (release 153), including 1,513 (19%) common SNVs”, and altogether “63% (5,057) of common SNVs identified in our cohort were either missing from dbSNP or are lacking accurate genotype information”.
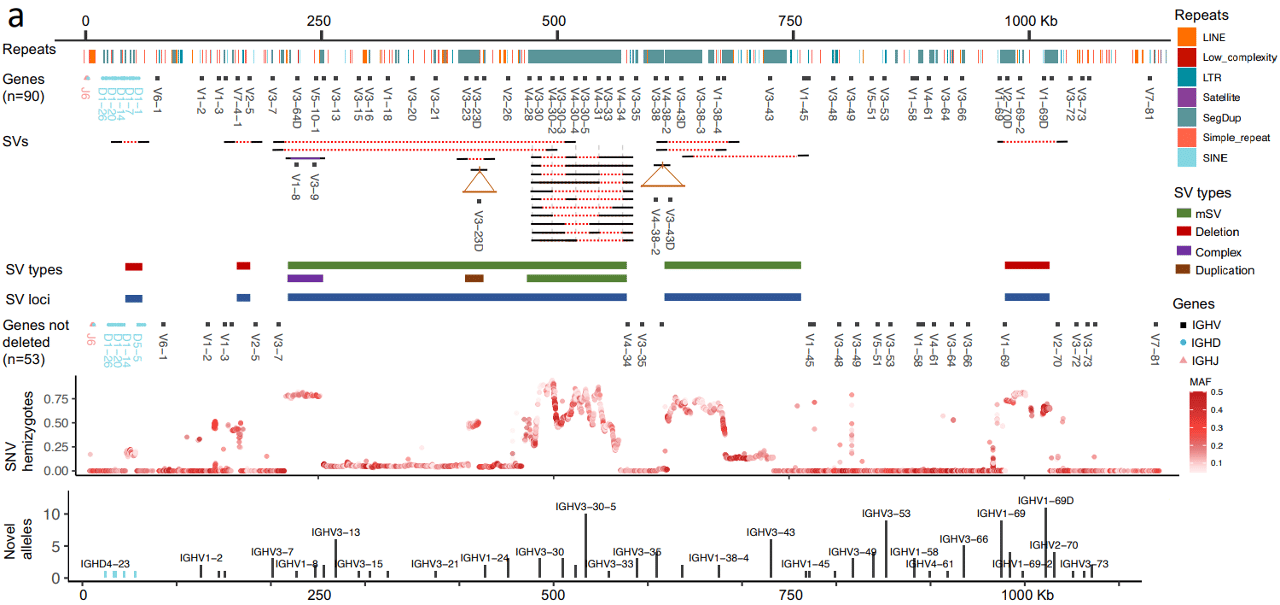
Ultimately, an improved understanding of antibody (Ab) repertoire diversity and function is the critical measure to better resolve the role of B cells in disease, so the next question the study addressed was whether this genetic diversity plays a role in an individual’s immune response, finding:
“Our results clearly demonstrate that genetics plays a significant role in shaping an individual’s antibody repertoire”
Because of the previous knowledge gaps, to date the models for immune responses and antibody repertoire dynamics have not accounted for genetic factors. Demonstrating here that “IGH genetic factors make significant contributions to gene usage in both the naive and antigen-experienced repertoire”, the study represents “a paradigm shift towards a model in which the Ab repertoire is formed by both deterministic and stochastic properties”, with “critical implications for delineating the function of Abs in disease”.
It follows that “these findings have the potential to reshape the way we conduct, analyze and interpret AIRR-seq [Adaptive Immune Receptor Repertoire sequencing] data, and use these data to profile the Ab response in disease”, and calls for the need to “more fully explore the extent of IGH polymorphism in the human population, as a means to resolve the role of germline variation in Ab function and disease.” It also extends to translational research, including better predictive models, diagnostic tools for disease and clinical phenotypes, improved designs and administration of therapeutics and vaccines, and “the ability to subset the population according to IG genotypes for more tailored healthcare decisions.”
Having done the heavy lifting on the IGH region, the researchers next made light work out of evaluating genetic variation in the IG Lambda (IGL) locus which plays crucial roles in antibody-antigen interaction and Ab structural stability. The preprint Characterization of the immunoglobulin lambda chain locus from diverse populations reveals extensive genetic variation paints a very similar picture – as with IGH, the complexity of the IGL locus has severely limited the effective use of short-read sequencing, limiting our knowledge of its population diversity. Using HiFi sequencing, the researchers generated haplotype-resolved assemblies for 16 individuals, identifying “significant allelic diversity, including 37 novel IGLV alleles”, and “highly elevated single nucleotide variation (SNV) in IGLV genes relative to IGL intergenic and genomic background SNV density”. Critically, when comparing SNV calls between these new datasets and existing short-read data, the authors “show a high propensity for false-positives in the short read datasets.” They conclude that the new HiFi-based resource represents “a significant advancement in our understanding of genetic variation and population diversity in the IGL locus.”
Understanding human immune responses, both at the population level as well as for a single individual, is central to advance precision medicine and to better human health. We are proud to partner with the scientific community in this endeavor, and are grateful for their application of HiFi sequencing to finally shed bright lights onto the most complex regions of the human genome, which are so critical for deciphering how humans respond to diseases.
Please connect with us to explore how PacBio HiFi sequencing can aid your human genomics research.
Read more about the HiFi Difference
Enabling the human pangenome reference
Enabling the era of reference pangenomes
More precise genomes for precision medicine
Not all gigabases are created equal