ACCURATE DETECTION OF CHALLENGING REPETITIVE DNA REGIONS WITH HIFI SEQUENCING
Leverage HiFi sequencing’s exceptional accuracy for precise characterization of tandem repeats, a prevalent form of structural variation in the human genome.
Repeat expansions can be challenging to sequence due to variability in length and high sequence homogeneity. Repeat loci can impact accuracy of read mapping, and short reads may have difficulty in aligning to regions of high similarity. Additionally, high GC content and the tendency of repetitive regions to form secondary structures can interfere with sequencing.
Sequencing repeat expansions is notoriously challenging due to their variable length, high sequence homogeneity, and complex GC-rich regions. These factors often lead to misidentification and inaccurate characterization by short-read sequencing, resulting in potential misrepresentation. PacBio long-read technology overcomes these obstacles, offering precise detection and a comprehensive view of repeat expansions, ensuring accurate insights where short-read sequencing falls short.
Harness the power of HiFi sequencing to characterize repeat expansions for:
- Understanding neurodegenerative disorders – Uncover the genetic variations linked to conditions such as Huntington’s disease and amyotrophic lateral sclerosis (ALS).
- Capturing variations underlying muscular dystrophy and ataxia – Detect key genetic markers that contribute to these disorders with extraordinary accuracy.
- Revealing genetic mechanisms of inherited conditions – Explore the genetic basis of inherited diseases through precise sequencing.
- Discovering novel de novo repetitive elements – Identify new and de novo repetitive sequences that could hold the key to understanding rare diseases.
PureTarget
Comprehensively genotype targeted repeat loci at scale to power neurological and carrier screening research.
Whole genome sequencing
Achieve more complete and phased human genome assemblies, providing deeper insights into the genetic complexity of health and disease.
Genetic testing
Illuminating complex regions of the genome that are crucial for disease identification.
Population genomics
Utilize scalable long-read sequencing to explore genomic variation, driving precision medicine initiatives in diverse populations.
Unlock new insights by profiling tandem repeats missed in standard whole genome analysis pipelines
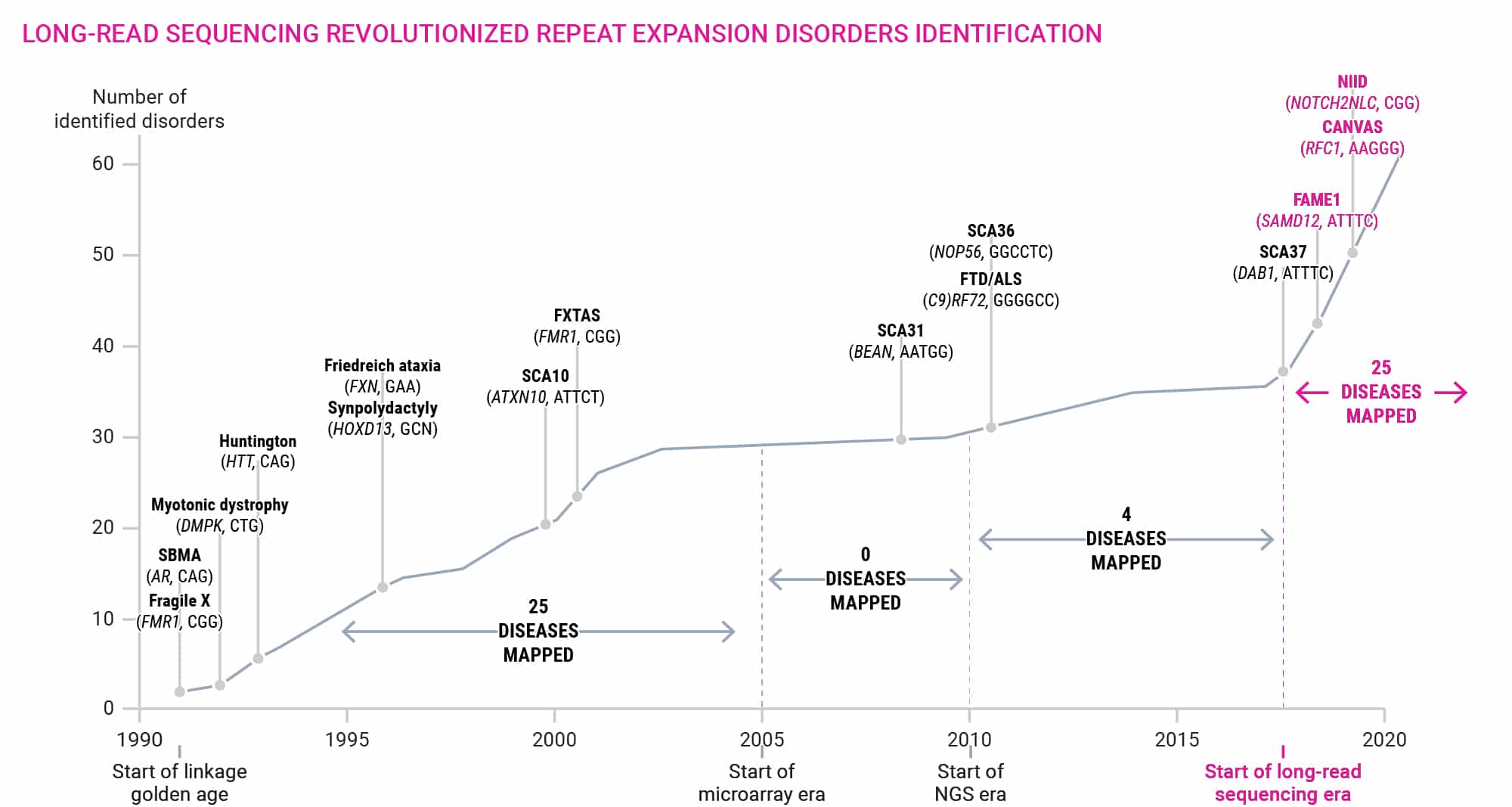
Tandem repeats are one of the most abundant types of variation in the human genome, and due to their repetitive nature, they have the highest mutation rate in the genome. This genomic instability is a major driver of disease. There are more than 50 disorders caused by expansions of short tandem repeats (STRs), and several variable number tandem repeats (VNTRs) have been shown to be involved in common complex disease like Alzheimer’s, autism, epilepsy, ALS, and others. Although tandem repeats play a key role in human health and disease, they are commonly not profiled as part of whole genome analysis pipelines.
This is not because they are considered unimportant, but because for so long it has been nearly impossible to sequence them using short-read sequencing, directly impacting analysis development. As part of the recent gapless human genome sequencing efforts, it has been estimated that 50–80% of structural variants are actually tandem repeats.8 However, since the current analysis pipelines are not optimized for tandem repeats, they are often either reported as indels if they are smaller or as clusters of variants if they are larger. HiFi sequencing enables a more complete and accurate calling of repeat expansions and identification of medically relevant interruption sequences along with methylation profiles.
This technology is crucial for screening individuals with these disorders to advance our understanding of human genetic diseases.
8Estimated by Adam English (Baylor College of Medicine) with Truvari software; SV from 36 haplotype-resolved assemblies spanning at least 50 bp; TR definitions from the UCSC SimpleRepeats track
9Depienne et. al (2021) 30 years of repeat expansion disorders: What have we learned and what are the remaining challenges? AJHG,108(5): 764-785
Blog
Sequencing 101: Tandem repeats
Delve into the challenges of sequencing tandem repeat loci and discover why HiFi sequencing is the optimal tool for both clinical and research characterization of these regions.
PureTarget repeat expansion panel spotlight
Target complex repetitive regions with PureTarget
Reliably and comprehensively capture 20 repeat expansion loci at scale, without the need for amplification, using the PureTarget repeat expansion panel.
Spotlight
HiFi sequencing identifies a common variant flanking a repeat loci associated with spinocerebellar ataxia, missed in reference genomes
Researchers from multiple institutions investigated the region around the FGF14 repeat locus, associated with spinocerebellar ataxia 27B, in over 2,000 PacBio whole genomes and multiple cohorts. The study found a common sequence variant flanking this region that “represents the major allele despite being absent from GrCh38.p14 and T2T-CMH13v2.0 assemblies”. Because HiFi sequencing reads are both long and highly accurate, this enables precise characterization of the length and variants within and around repeat loci, which can be crucial for understanding human disease.
Pellerin D, et al. (2024). A common flanking variant is associated with enhanced stability of the FGF14-SCA27B repeat locus. Nat Genet.
Blog
New hope for neurodegenerative diseases
Discover how Claire Clelland, MD, PhD leverages PureTarget amplification-free targeted sequencing of C9orf72 to further our understanding of dementia and other neurodegenerative diseases.
Application note
Tandem repeat analysis
Leverage computational tools with HiFi sequencing data for unprecedented detection and discovery of repeat expansions.
Repeat expansions in action
Resolving tandem repeats with TRGT
Characterization and visualization of tandem repeats at genome scale. Analyze tandem repeats in HiFi sequencing data with TRGT and visualize with TRVZ.


New methods to characterize VNTR variation in human genomes
Hear from Mark Chaisson, PhD on how HiFi sequencing helps accurately resolve VNTR regions.

Genome-wide catalog of de novo tandem repeats in rare disease trios
Learn more about TRGT-denovo, a tool for genome-wide mutation of de novo tandem repeats.

Accurately detect de novo tandem repeats
Identify all types of de novo tandem repeats to power your rare disease and population genomics studies.
Explore
Did you know we have a comprehensive library of reports, papers, and videos related to repeat expansions?




