EXPLORE MICROBIOME + METAGENOMICS WITH CONFIDENCE
Highly accurate long reads – HiFi reads – with single-molecule resolution are ideal for full-length 16S rRNA sequencing, shotgun metagenomic profiling, and metagenome assembly, so that you can:
- Determine community composition at the species or strain-level with competitively priced full-length 16S sequencing
- Identify 6-8 full-length genes in every HiFi read with efficient, cost-effective metagenomic profiling
- Generate hundreds of high-quality (HQ) metagenome assembled genomes (MAGs), many of which are single contig, circular MAGs
- Leverage epigenomic data to associate contigs and plasmids from closely related strains
Brochure
メタゲノムとマイクロバイオームシーケンスの新たなスタンダードを構築
メタゲノムおよびマイクロバイオームシーケンスへ高精度で柔軟なHiFiソリューションを用いることで、研究者は培養の必要なく高解像度で微生物や微生物群集を研究することができます。HiFiシーケンスの優れた精度と長いリード長により、科学者が必要とする柔軟性と高解像度を実現します。
MICROBIAL SEQUENCING METHODS
HIFI METAGENOMICS SEES PERFORMANCE ENHANCEMENTS ACROSS THE BOARD
Shotgun metagenomics with PacBio highly accurate long-read sequencing (aka HiFi sequencing) has been the premier approach for generating microbial datasets that are both very high quality and stunningly complete. Now, the Revio system offers faster and more cost-effective long-read sequencing for metagenomics researchers, allowing for more high-quality and complete microbial datasets and access to new frontiers of discovery.
Spotlight
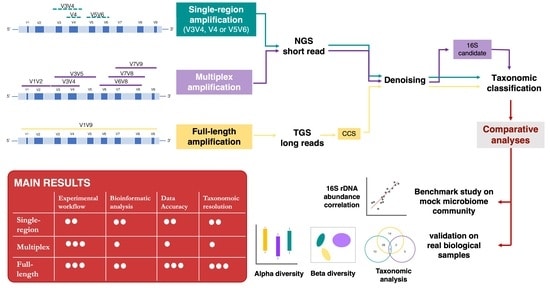
WHY GUESS WITH PARTIAL 16S SEQUENCING WHEN FULL-LENGTH 16S IS THE BEST?
This study compared three different methods to achieve the deepest microbiome taxonomic characterization: (a) the single-region approach, (b) the multiplex approach, covering several regions of the target gene/region, both based on NGS short reads, and (c) the full-length approach, which analyzes the whole length of the target gene thanks to TGS long reads [PacBio]. Analyses carried out on benchmark microbiome samples, with a known taxonomic composition, highlighted a different classification performance, strongly associated with the type of hypervariable regions and the coverage of the target gene. Indeed, the full-length approach showed the greatest discriminating power, up to species level, also on complex real samples. This study supports the transition from NGS to TGS [PacBio] for the study of the microbiome.
Notario, E., et al. (2023). Amplicon-Based Microbiome Profiling: From Second- to Third-Generation Sequencing for Higher Taxonomic Resolution. Genes, 14(8), 1567. https://pubmed.ncbi.nlm.nih.gov/37628619/
Spotlight
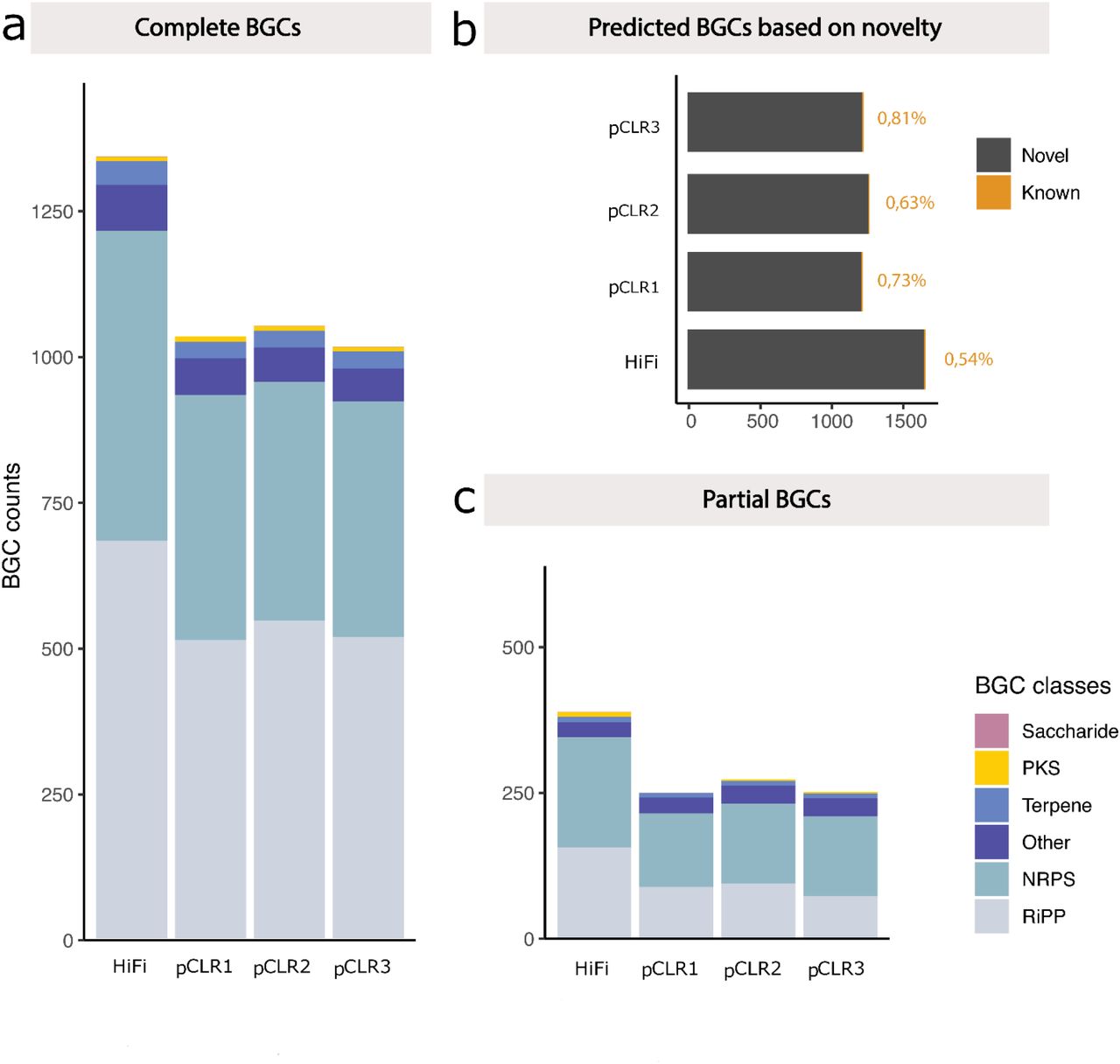
メタゲノム解析と機能プロファイリングには、リードの長さと精度が重要
羊の糞便サンプルのHiFiデータを用いた解析結果と、同じデータセットから精度の低いサブリードを用いた結果を比較すると、精度向上によるアセンブリの連続性、MAG回収、遺伝子発見へのインパクトがわかります。左の図では、より連続性の高いHiFiアセンブリにより、ノイズの多いCLRロングリードよりも25%多く完全な生合成遺伝子クラスタ(biosynthetic gene clusters : BCG)を回収することができました。
Bickhart, D.M. et al. (2022). Generation of lineage-resolved, complete metagenome-assembled genomes from complex microbial communities.. Nature Biotechnology, 0.1038/s41587-021-01130-z
Application Brief
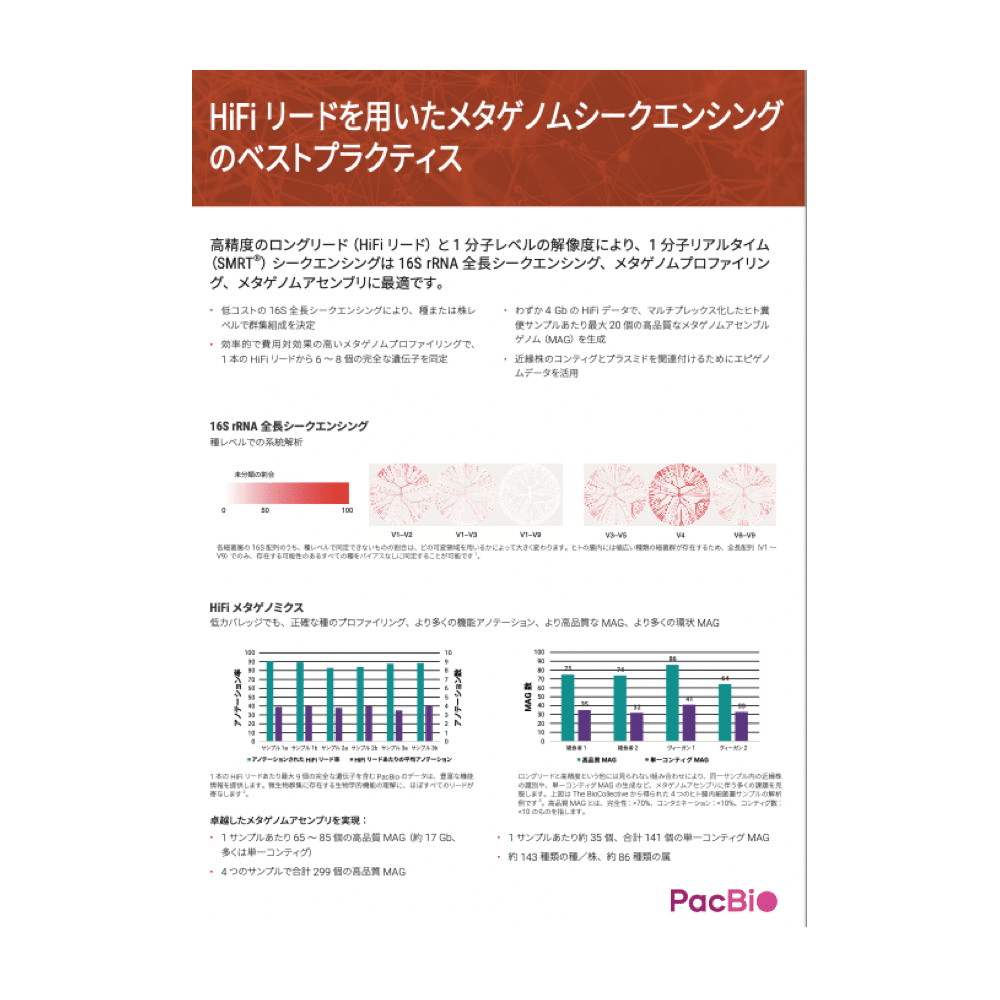
HIFI リードによるメタゲノムシーケンス
HiFiリードを用いたメタゲノム解析のベストプラクティスをご紹介
高精度のロングリード(HiFi リード)と1 分子レベルの解像度により、1 分子リアルタイム(SMRT®)シークエンシングは16S rRNA 全長シークエンシング、メタゲノムプロファイリング、メタゲノムアセンブリに最適です。
PacBioに問い合わせる