Resolve complex genomics underlying human disease
Single Molecule, Real-Time (SMRT) sequencing offers a flexible solution for targeting regions of interest, from several hundred base pairs to kilobase scale, and delivers the most comprehensive view of the associated genes. With highly accurate long reads (99.9% single-molecule read accuracy) it delivers the read length and accuracy needed to improve fine mapping and simplify the analysis of complex portions of the genome. SMRT sequencing gives you the ability to:
- Rapidly screen and identify all variants
- Discover haplotype-specific markers
- Resolve difficult-to-sequence regions, regardless of size
- Confirm insertion sites of transgenes and validate gene editing events
Spotlight
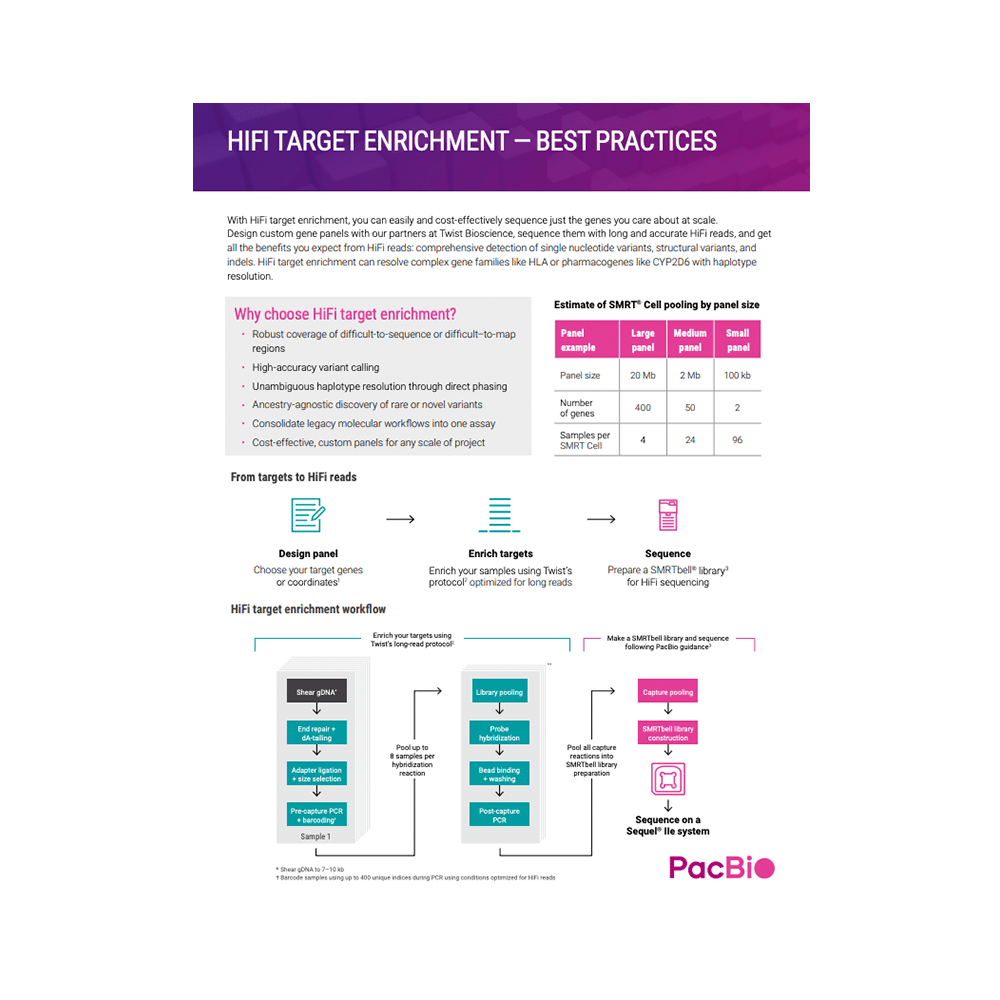
HIFI TARGET ENRICHMENT WITH TWIST PROBES
Learn how you can get all the benefits of long and accurate HiFi reads at scale.
Sample and library prep
Fast and easy library preparation performed in a single tube.
- Start with high-quality, nucleic acids, as low as 250 ng for a 250 bp amplicon
- Create SMRTbell templates of amplicons of various sizes, from hundreds of bases to kilobase scale.
- Optimize throughput with flexible barcoding options
- Amplify targets with one round of PCR using target-specific primers with incorporated barcodes
- Add barcodes during PCR using a two round design with PacBio’s barcoded M13 primer plate
- Add barcodes during SMRTbell library prep with PacBio’s barcoded SMRTbell adapter plate
- Multiplex up to 10,000 samples per SMRT Cell
Sequencing
Use the Sequel systems to accurately sequence targets in the human genome.
- Generate HiFi reads
- Q20 single-molecule accuracy reads
- Up to 4,000,000 reads per SMRT Cell 8M
- Sequence to desired coverage based on project needs
- Target 30-fold coverage for variant detection
- Increase coverage for minor variant detection (~6,000-fold coverage for 1% sensitivity)
Data analysis
Analysis solutions for every user in the lab with the PacBio analytical portfolio.
- Fully characterize genetic complexity — structural variation, rare SNPs, indels, CNV, microsatellites, haplotypes, and phasing
- Utilize a variety of analysis tools within SMRT Link
- Generate HiFi reads (>Q20) using circular consensus sequencing (CCS) mode
- Easily demultiplex barcodes within SMRT Link
- Detect, quantitate, and phase single nucleotide polymorphisms within coding regions using minor variants analysis
- Output data in standard file formats (BAM and FASTA/Q) for seamless integration with downstream analysis tools
- Perform reference-free analysis of amplicons with pb amplicon analysis (pbaa)
- HiFi reads are compatible with standard analysis tools for variant calling such as GATK and Deep Variant.
Application note
HiFi amplicon sequencing for pharmacogenetics: CYP2D6
With PacBio® HiFi sequencing, CYP2D6 variation can be captured using highly accurate long reads on a single platform. Amplicon-based HiFi sequencing fully and directly resolves and phases complex loci like CYP2D6 without assembly or inference, allowing for ancestry-agnostic haplotype assignment. In this application note, we propose a streamlined PCR amplicon workflow that characterizes full-length CYP2D6 alleles with PacBio HiFi sequencing.
Application note
Targeted HiFi sequencing for congenital adrenal hyperplasia
In this application note, we focus on HiFi sequencing of full-length PCR amplicons spanning the 3.3 kb CYP21A2 and its pseudogene, CYP21A1P. PacBio HiFi reads are long (up to 25 kb) and accurate (99.9%), making them well suited to analyze the complexity of CAH-related pathogenic variants, where gene conversions and chimeric genes are the predominant mutation types.
Application note
Targeted HiFi sequencing for Thalassemia
PacBio HiFi reads are long (up to 25 kb) and accurate (99.9%). Targeted HiFi sequencing of amplicons has the ability to span entire genes, allowing for straightforward haplotype construction, detection of structural variants or copy number variants in addition to SNVs and indels. These attributes make targeted HiFi sequencing well suited to analyze both rare and common variants of all types, thus increasing the positive detection rate of thalassemia-related variants.
Spotlight
SMRT Sequencing uncovers novel causative mutations and resolves duplicated genes
PacBio long-read sequencing enabled scientists to detect a single causative mutation for autosomal dominate tubulointerstitial kidney disease (ADTKD) in MUC1 gene residing in a GC-rich variable number tandem repeat region (VNTR) that was previously intractable. Explore this research further:
Wenzel, A. et al., 2018. Single molecule real time sequencing in ADTKD-MUC1 allows complete assembly of the VNTR and exact positioning of causative mutations. Scientific Reports, 8(1), p.4170.
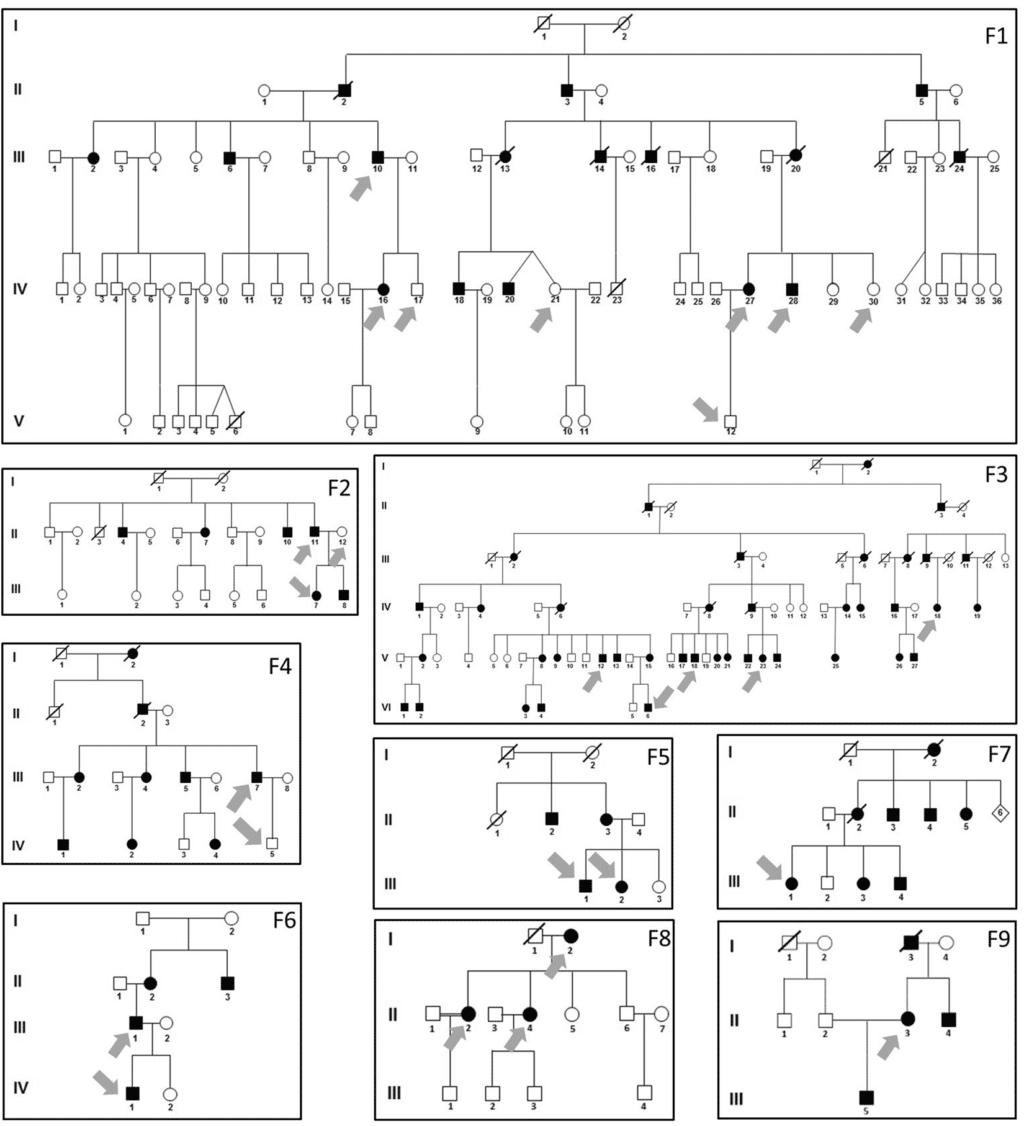
Application brief
Targeted sequencing for amplicons best practices
Learn more about these best practices for targeted sequencing of amplicons.