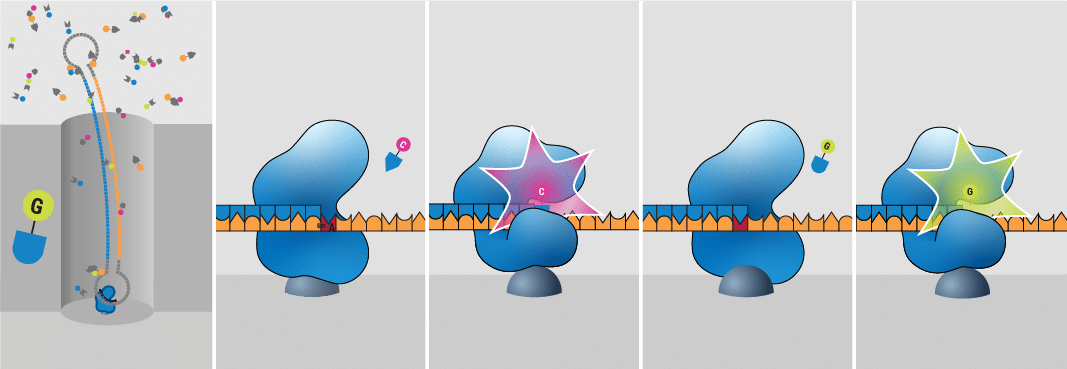
How does HiFi sequencing work?
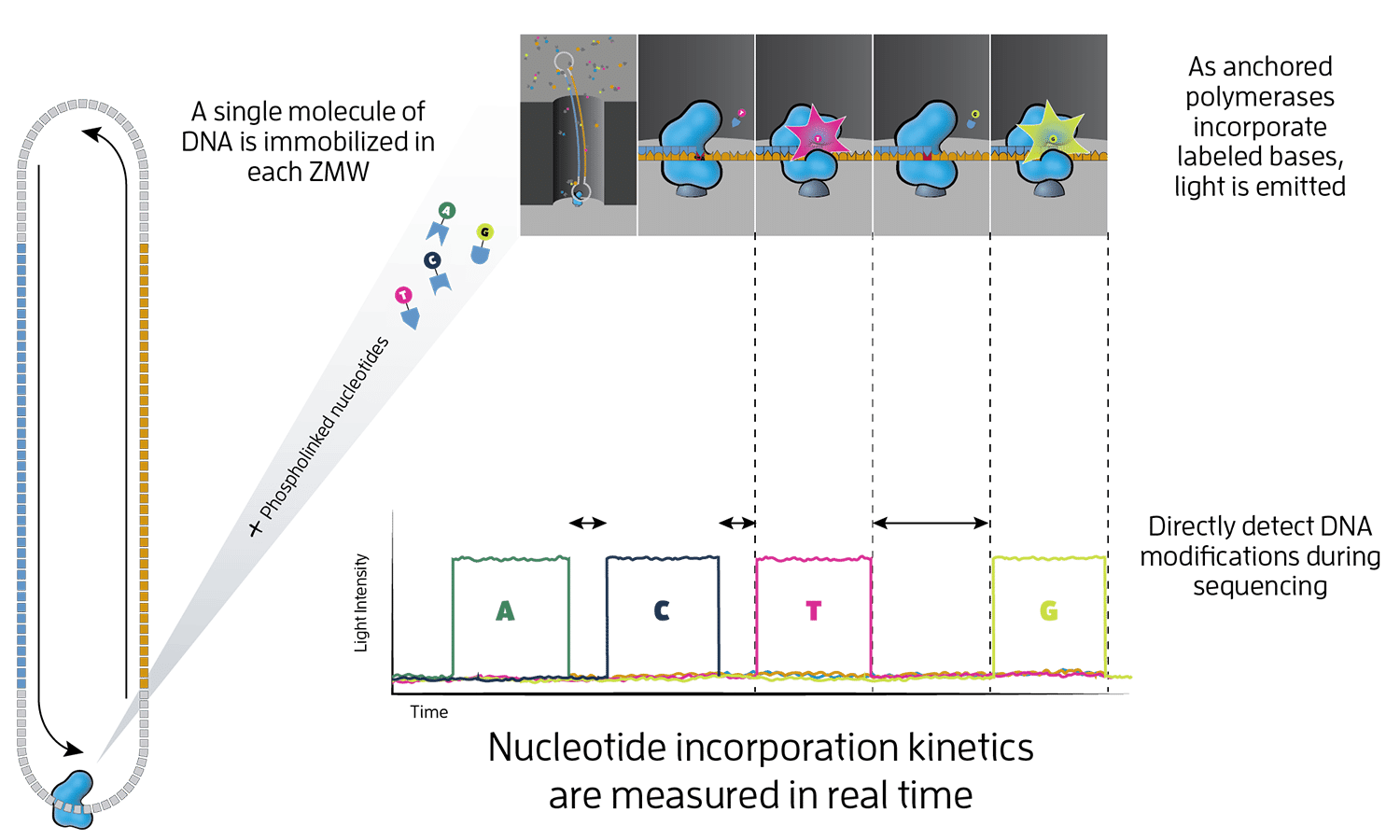
HiFi sequencing is a single-molecule, real-time sequencing technology (SMRT) that provides incredible single-molecule read accuracy across long reads of tens of kilobases in length or more. HiFi reads are generated by combining information from multiple observations of a single DNA molecule, resulting in over 99% accuracy of individual HiFi reads.
Developed by PacBio, this process occurs inside small wells on a special microchip called a SMRT Cell, which contains millions of these tiny wells. HiFi sequencing uses fluorescent light signals to identify DNA bases and modified bases (without bisulfite treatment). As a polymerase enzyme adds new nucleotide bases to a newly replicated strand, it emits tiny flashes of light. This technology powers our HiFi long-read sequencing platforms, providing reliable long reads and accurate results.
From DNA to discovery, learn about the steps of a HiFi sequencing workflow.
Long-read sequencing myths: debunked
What are the advantages of HiFi sequencing?
Long reads
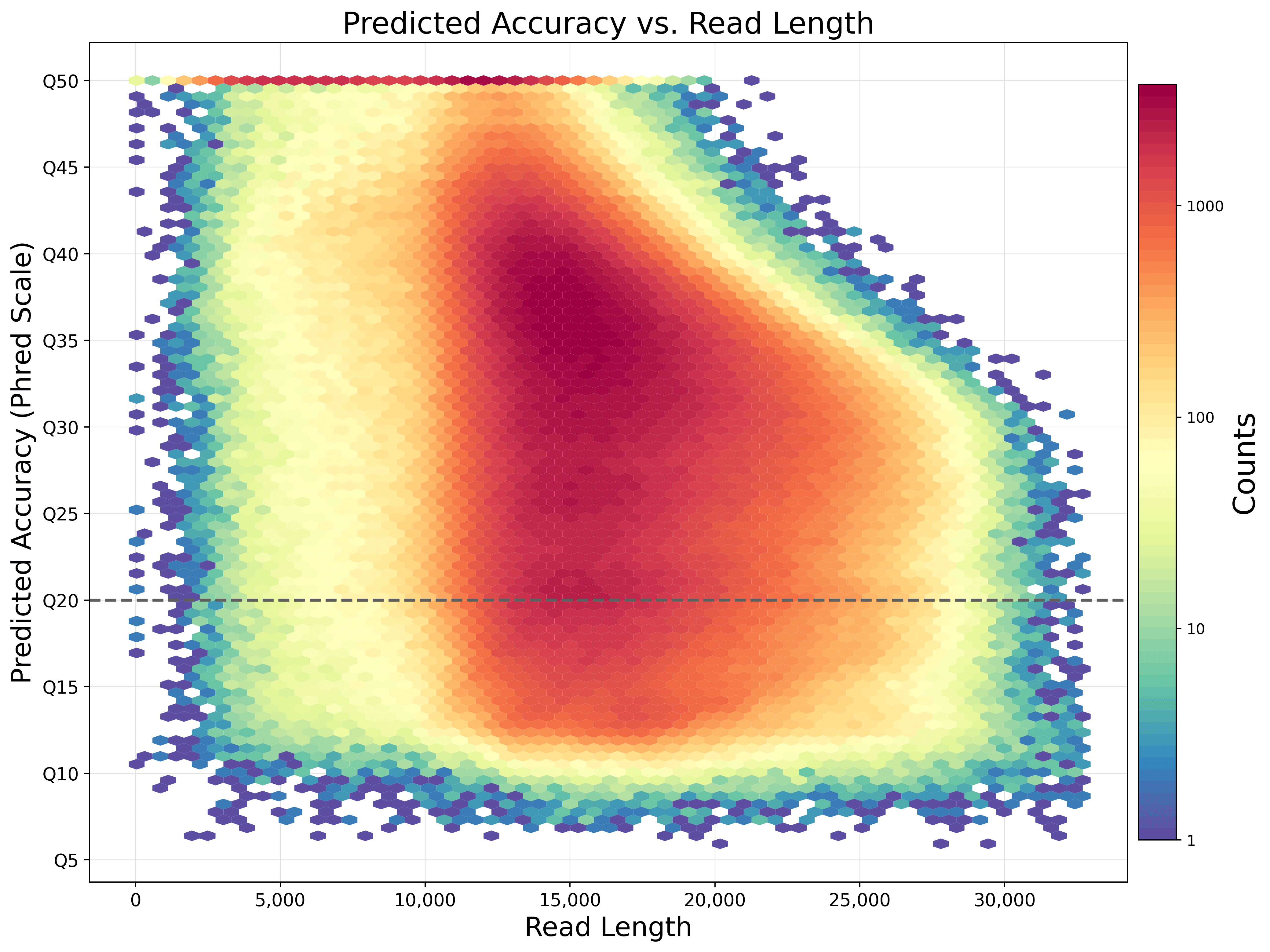
With reads tens of kilobases in length you can readily assemble complete genomes and sequence full-length transcripts. HiFi sequencing provides exceptional read lengths without compromising throughput or accuracy.
Data from an HG002 human library using the SMRTbell express template prep kit 2.0 on a Sequel IIe system (2.0 chemistry, Sequel IIe system software v10, 30-hour movie). Read lengths, reads/data per SMRT Cell, and other sequencing performance results vary based on sample quality/type and insert size.
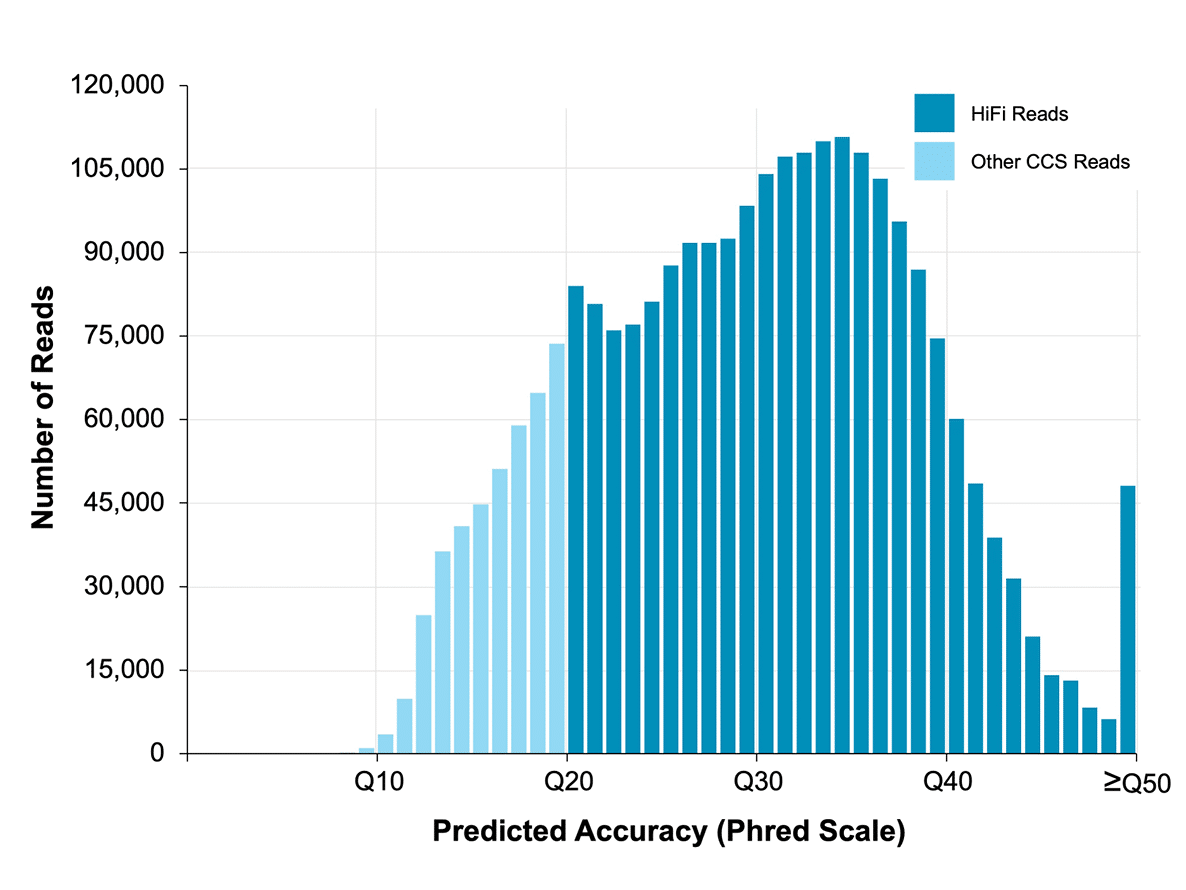
High accuracy
Sequencing free of systematic error achieves 99.9% read accuracy and >90% of bases Q30+. Explore the benefits of highly accurate long-read sequencing. With exceptional accuracy, low sequencing-context bias, and accurate mapping of reads, HiFi sequencing provides the information needed to confidently call and detect all variants.
Data from a 15 kb size-selected human library using the SMRTbell express template prep kit 2.0 on a Sequel IIe system (2.0 chemistry, Sequel IIe system software v10, 30-hour movie). Read lengths, reads/data per SMRT Cell, and other sequencing performance results vary based on sample quality/type and insert size.
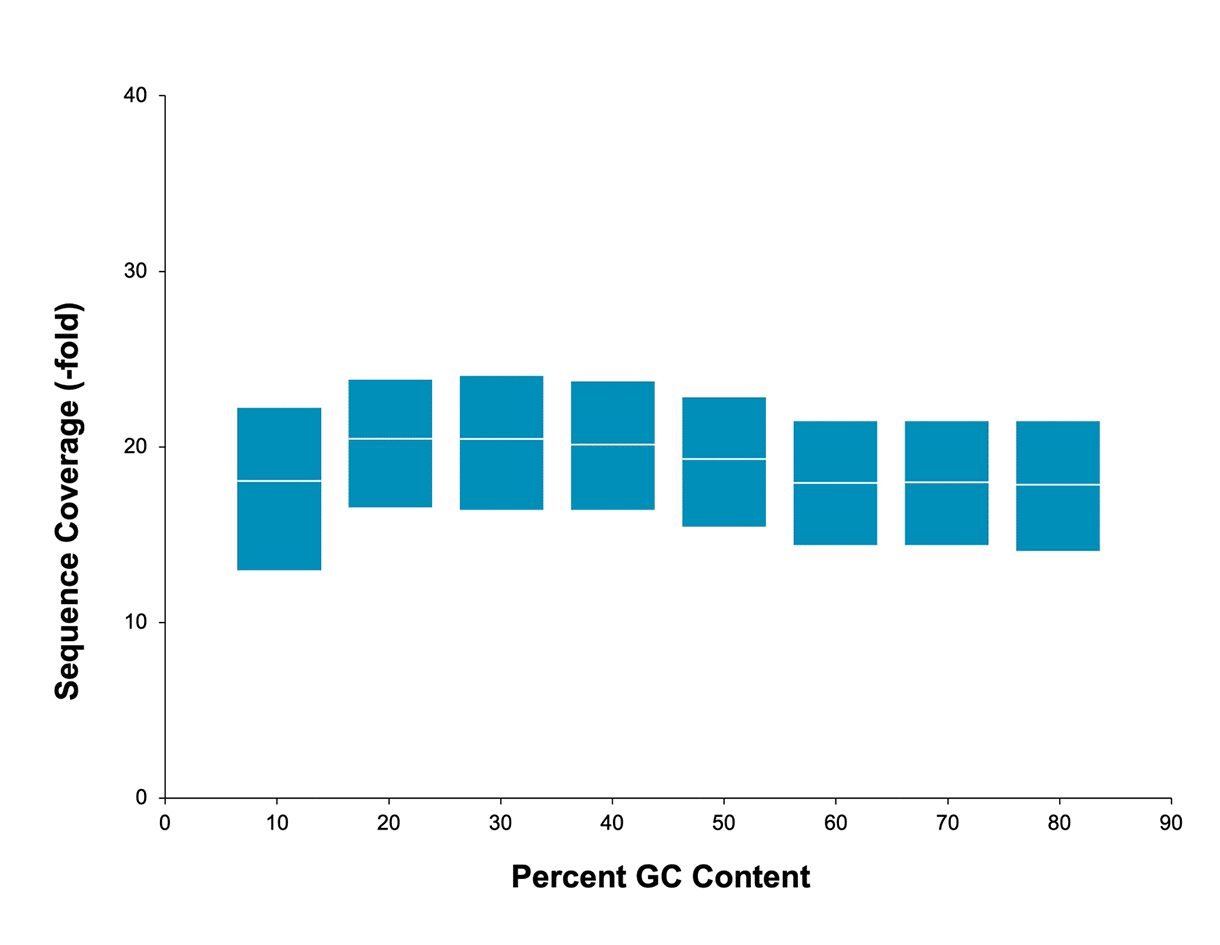
Uniform coverage
No bias based on GC content means you can sequence through regions inaccessible to other technologies. Readily sequence through AT-rich or GC-rich regions, highly repetitive sequences, long homopolymers, and palindromic sequences with HiFi sequencing.
Mean coverage per GC window across a human sample. Data generated with a 20 kb HiFi library on a Sequel II system (2.0 chemistry and Sequel II system). Read lengths, reads/data per SMRT Cell, and other sequencing performance results vary based on sample quality/type and insert size.
Native molecules
Capturing sequence data from native DNA or RNA molecules enables highly accurate long reads with >99.9% single-molecule accuracy. Readily achieve high-quality reads to confidently resolve variants of all types.
Data from a 15 kb size-selected human library using the SMRTbell express template prep kit 2.0 on a Sequel IIe system (2.0 chemistry, Sequel IIe system software v10, 30-hour movie). Read lengths, reads/data per SMRT Cell, and other sequencing performance results vary based on sample quality/type and insert size.
Epigenetics
With no PCR amplification step, base modifications are directly detected during sequencing. Measurement of variation in polymerase kinetics of DNA base incorporation eliminates the need for chemical modification to detect base modifications. This allows you to capture sequence and epigenetic information in a single experiment. See how on-instrument 5-base HiFi sequencing detects methylation without additional library preparations.