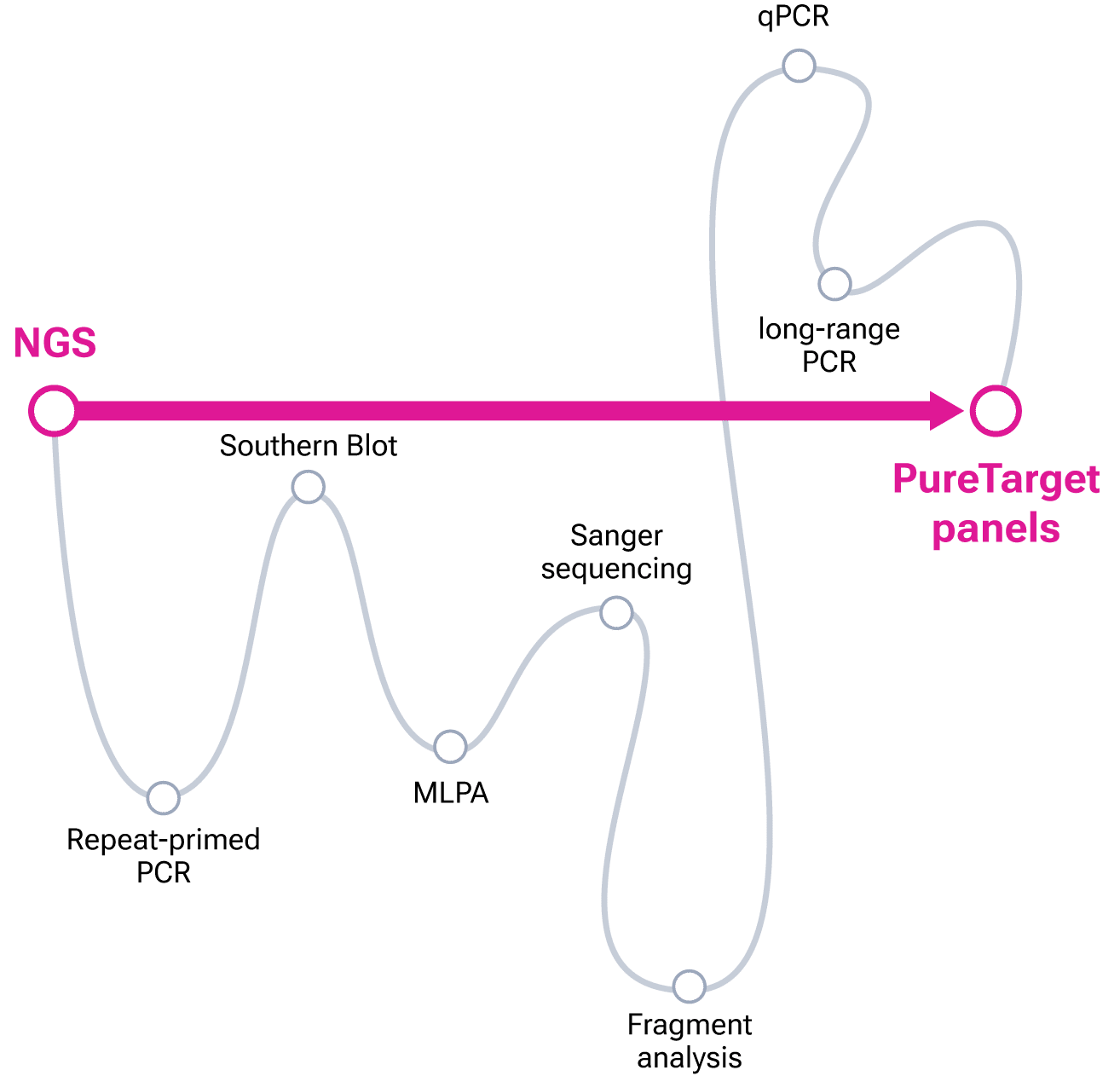
Consolidate legacy methods into a single assay with 1-day PureTarget prep on the Vega system
Modernize your molecular assays with the PureTarget repeat expansion panel on the Vega benchtop system.This highly accurate long-read sequencing panel lets you consolidate multiple methods with a 1-day prep, 48-sample multiplexing, and the preservation of crucial methylation data. As an amplification-free prep, PureTarget allows you to easily resolve challenging repeats in the FMR1, C9orf72, and RFC1 genes which short reads can’t fully capture. This benchtop panel keeps you in control, getting fast results without relying on third-party sequencing services.
Highlights:
Key metrics

Comprehensive genotyping of the most challenging targets
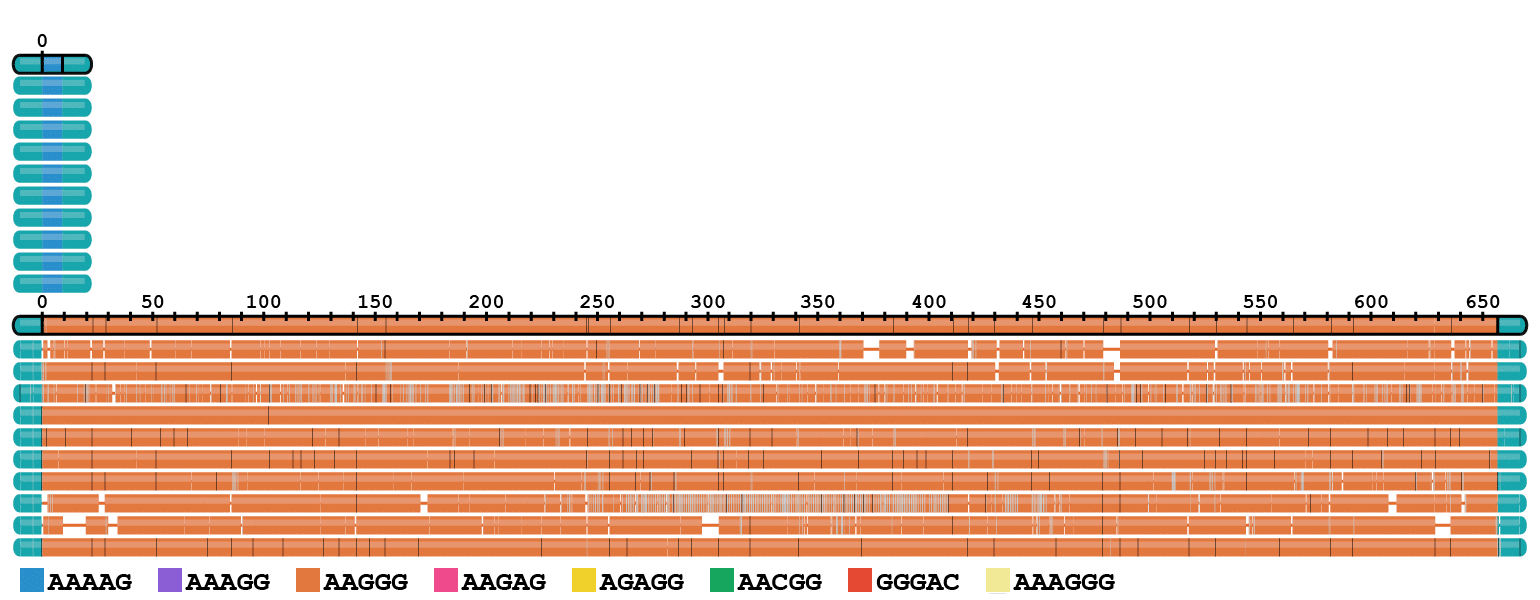
Repeat expansions in the FXN and RFC1 genes occur in high frequency across many human populations, with recent estimates that 1 in ~14 individuals carry pre–mutations at RFC1 and 1 in ~100 carry pre-mutations at FXN (Ibañez et al., 2024). Accurate repeat sizing is important to identify carriers of pathogenic alleles longer than 70 repeats for FXN and longer than 400 repeats at RFC1 (Leitão et al., 2024). Long and accurate HiFi reads make it possible to distinguish among the diverse repeat motifs at RFC1, only some of which are pathogenic (Dominik et al., 2023).
RFC1 expansions
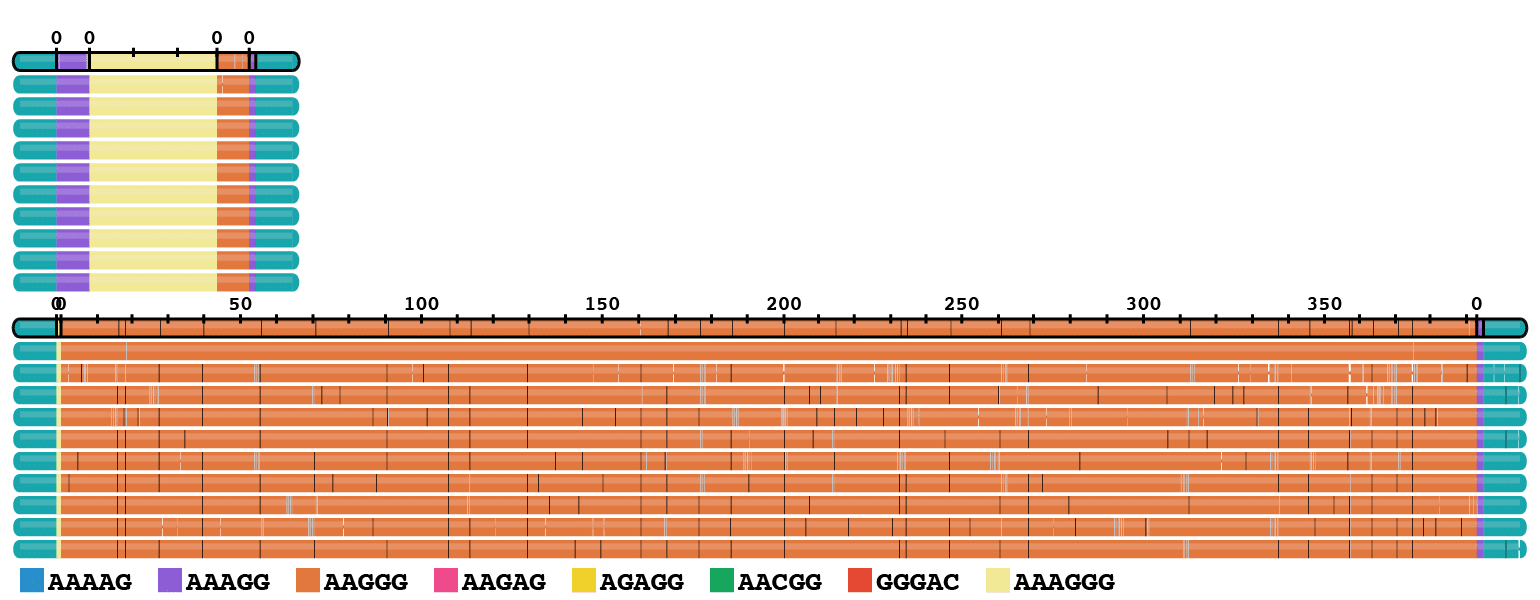
FXN expansions
| Long allele | Short allele | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Coverage | Observed motif count | Expected motif count | Coverage | Observed motif count | Expected motif count | ||||
| HM16212 | 92 | 471 | 500 | 146 | 8 | <30 | ||||
| NA16212 | 68 | 515 | 500 | 99 | 8 | <30 | ||||
| NA16202 | 49 | 817 | 830 | 86 | 8 | <30 | ||||
| NA16237 | 49 | 699 | 700 | 134 | 8 | <30 | ||||
High on-target coverage enables accurate genotyping
The PureTarget repeat expansion panel enables deep coverage for accurate genotyping of repeat expansions associated with human disease. The table below highlights typical coverage for Coriell samples with known repeat expansions. Note that 2 µg of DNA was prepared with the PureTarget repeat expansion panel of 20 targets, sequenced on the Vega system, and analyzed with TRGT (Dolzhenko et al., 2024) in the SMRT Link PureTarget repeat expansion panel analysis workflow. Full dataset available here.
| Sample | Target with repeat expansion | Total reads per sample | Mean target coverage | Coverage of expanded allele | Epanded allele size (bp) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 20 gene panel | ||||||||||
| HM23709 | AR | 54,252 | 343 | 182 | 147 | |||||
| NA13716 | ATN1 | 40,700 | 234 | 88 | 216 | |||||
| NA13536 | ATXN1 | 39,845 | 267 | 132 | 124 | |||||
| NA06153 | ATXN3 | 64,164 | 258 | 114 | 207 | |||||
| ND14442 | C9orf72 | 16,821 | 51 | 4 | 4563 | |||||
| HM03756 | DMPK | 52,504 | 395 | 282 | 1305 | |||||
| NA09237 | FMR1 | 36,167 | 257 | 113 | 2683 | |||||
| NA16215 | FXN | 67,939 | 245 | 62 | 2963 | |||||
| NA13509 | HTT | 58,870 | 480 | 225 | 252 | |||||
| HM23629 | PABPN1 | 50,569 | 417 | 218 | 27 | |||||
| HG01175 | RFC1 | 53,454 | 378 | 57 | 1948 | |||||
3 days from sample to insight
Extraction
Start with HMW DNA extracted from human blood or cell lines using Nanobind extraction kits.
Library prep
Targeting and library prep can be completed in 8 hours. All reagents supplied in the kit.
Sequencing
Sequence up to 48 samples per Vega SMRT Cell with 24 hour movies and optimized run conditions for PureTarget libraries.
Data analysis
SMRT Analysis is automatically configured to generate library QC, target enrichment statistics, and tandem repeat genotypes using TRGT (Dolzhenko et al., 2024).
References
Dolzhenko, E., et al. (2024). Characterization and visualization of tandem repeats at genome scale. Nat Biotechnol. 2024 doi: 10.1038/s41587-023-02057-3.
Dominik, N., et al. (2023). Normal and pathogenic variation of RFC1 repeat expansions: implications for clinical diagnosis. Brain. 146(12):5060-5069. doi: 10.1093/brain/awad240.
Ibañez, K., et al. (2024) Increased frequency of repeat expansion mutations across different populations. Nature Medicine. https://doi.org/10.1038/s41591-024-03190-5
Leitão, E., et al. (2024). Identification and characterization of repeat expansions in neurological disorders: Methodologies, tools, and strategies. Rev Neurol (Paris). 180(5):383-392. doi: 10.1016/j.neurol.2024.03.005.
Explore other Vega system datasets
Now, you can
Vega makes high-accuracy sequencing accessible to labs of all sizes, because every lab deserves the power of HiFi.

