Long-read PacBio HiFi sequencing has made transformative contributions to many areas of genomics over the past few years, including de novo assembly, calling of all variant types, full-length phased targeted and whole genome sequencing, full-length RNA sequencing, and high-resolution metagenomics. Underlying these contributions is the highly accurate, end-to-end sequencing of native long DNA molecules. Over the past decade, attempts have been made to synthetically reconstruct longer-molecule sequences from short sequence reads using short-read technologies, typically involving copying the original molecule using PCR. Examples such as Moleculo or 10X Genomics linked reads have proven cumbersome in application – for sample prep and analysis – and were subsequently abandoned. Recently, Illumina announced ‘Infinity’ reads and Element Biosciences acquired Loop Genomics as the latest iteration of this concept of synthetic long reads.
While little public data is available for either of the new synthetic long read approaches, Illumina showed an example comparison earlier this year at the Festival of Genomics & Biodata conference (FoG 2022). In the IGV screenshot presented (below), synthetic Infinity reads – labeled “Longas” – are at the top, followed by standard Illumina short reads, and PacBio HiFi reads labeled “CCS” depicted at the bottom:
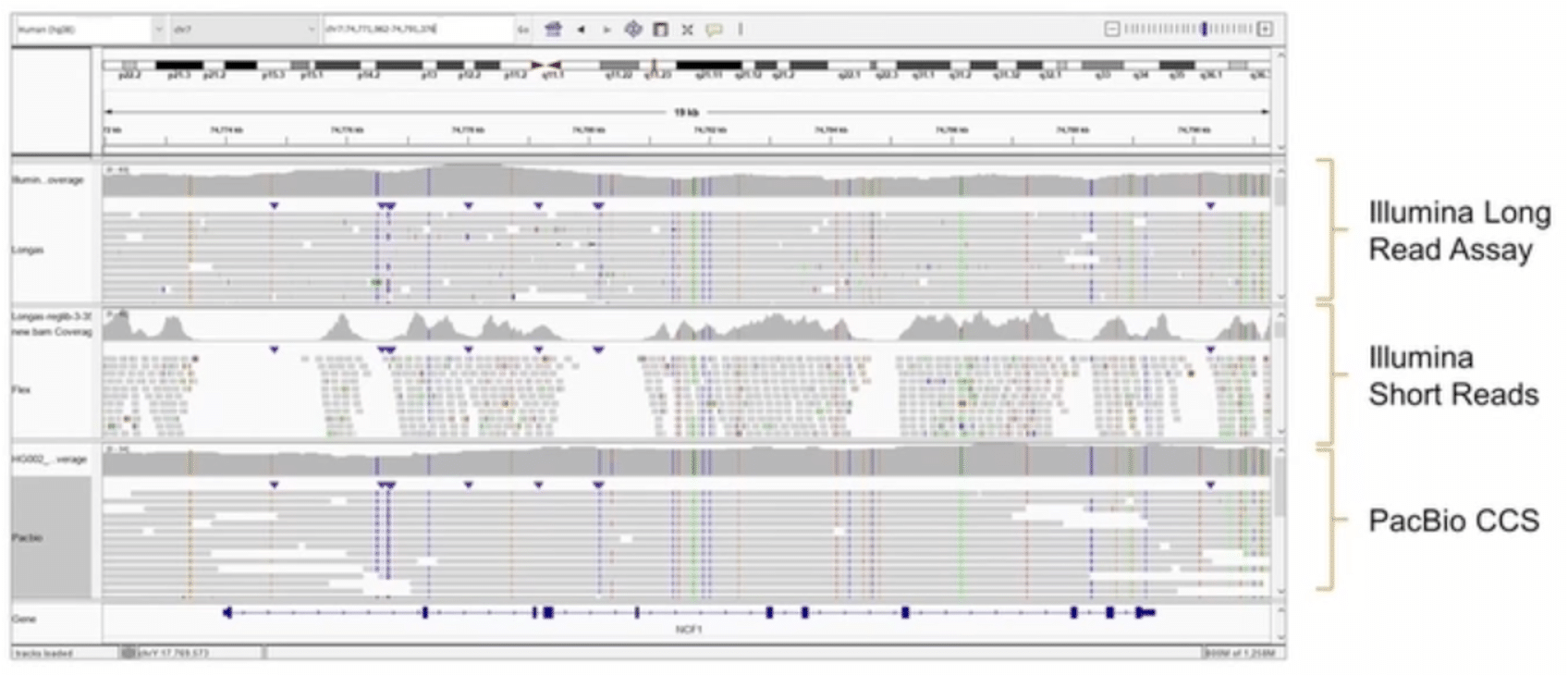
However, a close comparison highlights important advantages of PacBio HiFi reads (Figure 2), including:
a) Variant calling accuracy: A variant (a tandem repeat insertion), clearly resolved as homozygous with PacBio HiFi reads, is only partially detected with Infinity, making the variant falsely appear as heterozygous.
b) Variant calling confidence: A region of two adjacent homozygous and heterozygous SNVs, which are clearly resolved with PacBio HiFi reads, are indicated in the Illumina Infinity coverage panel, but poorly supported by the underlying reads.
c) Long-range phasing: the IGV region depicted contains heterozygous variants, separated by ~10 kb, which are clearly phased through several PacBio HiFi reads that fully span this distance, thereby allowing direct phasing (as cis of the two variants relative to the reference). Illumina Infinity reads are not sufficiently long to span the distance between the two variants.
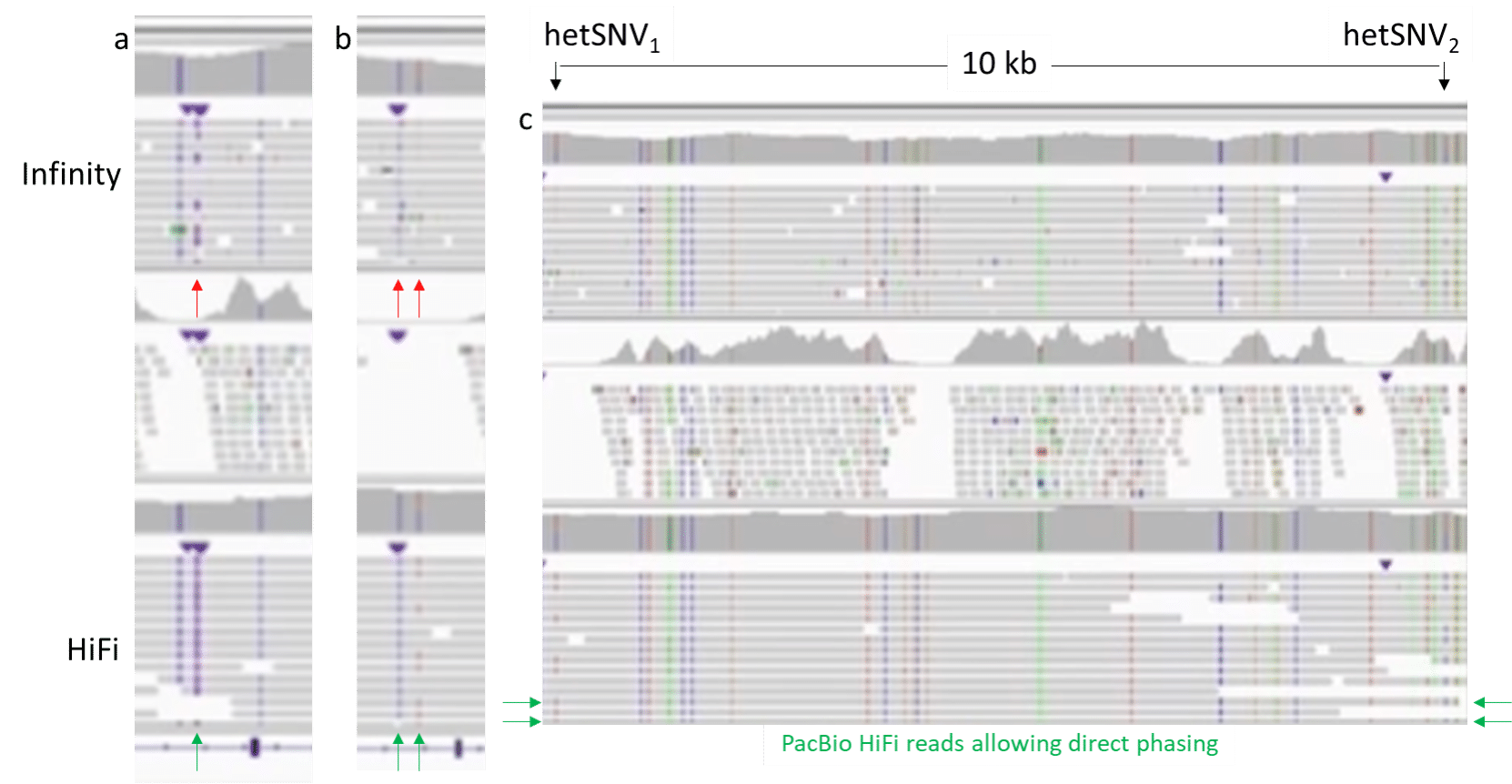
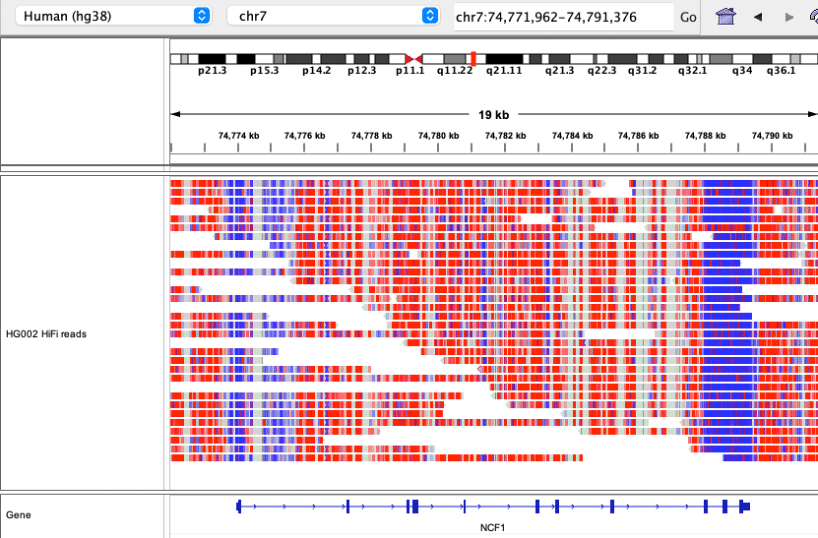
Further, true long reads directly detect CpG methylation, which short reads do not nor is it possible with the PCR used for the synthetic read approaches.
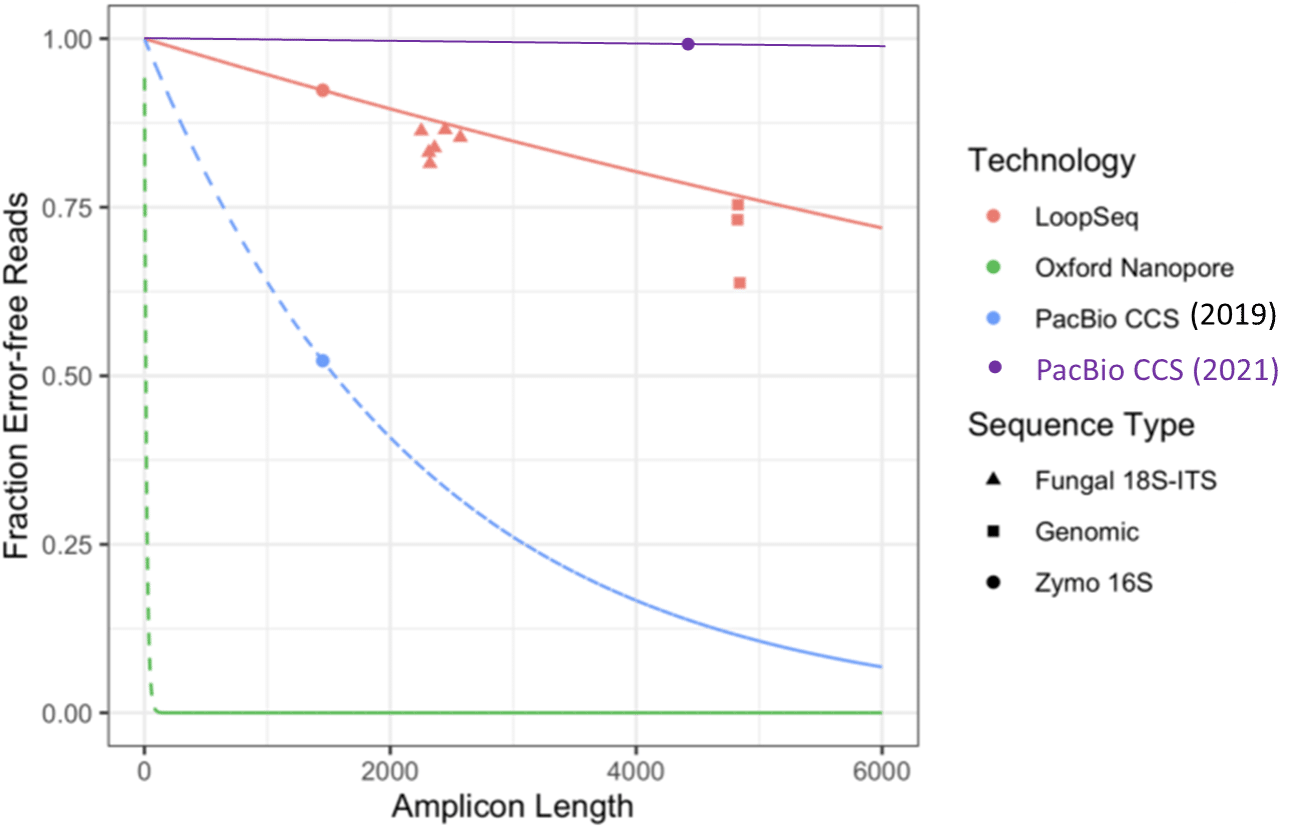
This week, Element Biosciences described synthetic long reads using Loop Genomics technology, and showed a technology comparison graph for metagenomic amplicon sequencing (Figure 3 from Callahan et al. (2021)):
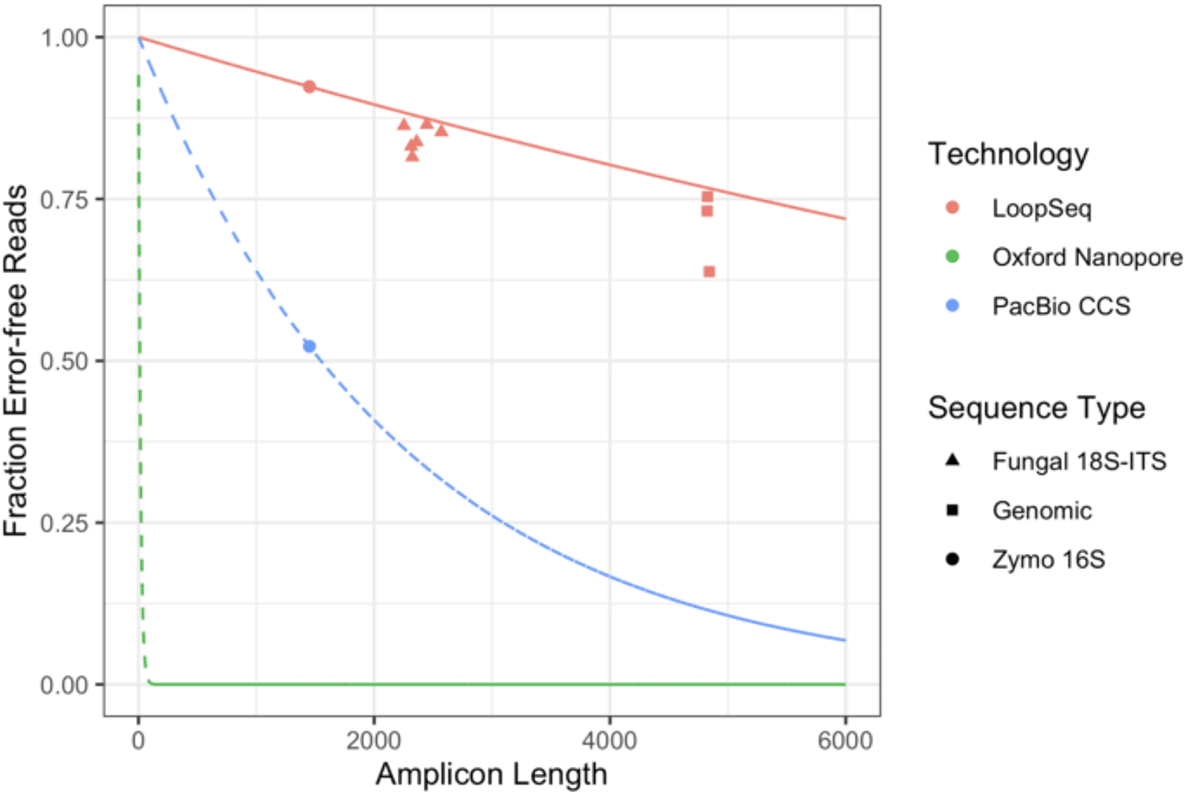
The “PacBio CCS” datapoint was taken from a 2019 publication that does not represent today’s performance. For example, Karst et al. (2021) described HiFi sequencing of full-length (4.4 kb) rDNA amplicons of the same sample at 99.9993% (Q51.5) accuracy, almost two orders of magnitude greater than the previous study. Adding this datapoint to the graph illustrates how the superior accuracy of HiFi sequencing results in better application performance:
These examples re-affirm what the scientific community has concluded for years – synthetic long reads do not compare to the benefits of true long reads produced by PacBio HiFi sequencing. Even further, synthetic long reads run the risk of confounding studies with artifacts that can provide potentially inaccurate information.
PacBio HiFi sequencing provides the most accurate, contiguous, and complete genomic and epigenomic information with ever increasing applications to enable the promise of genomics to better human health.
Read more about the HiFi Difference
More precise genomes for precision medicine
Not all gigabases are created equal